Get involved
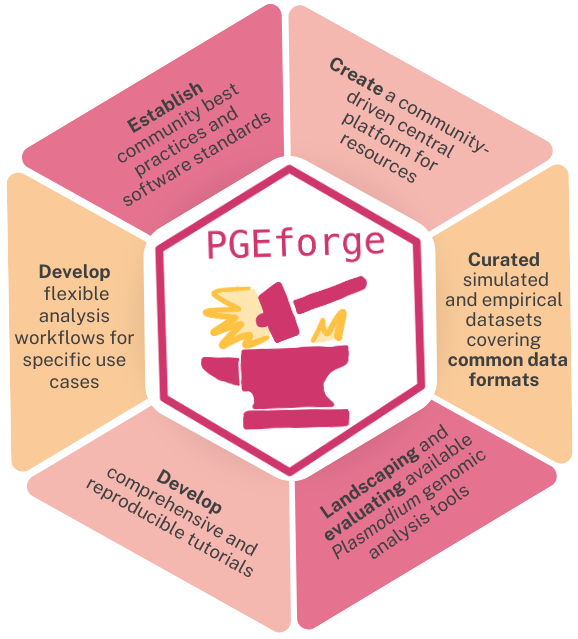
The PGEforge vision
PGEforge is a community-driven platform created by and for malaria genomic epidemiology analysis tool developers and end-users. If you are interested in contributing to these efforts, please get involved!
How to contribute to PGEforge
We are excited to have you join us and we welcome input from all areas of the research community to continuously improve and expand our platform. PGEforge aims to contribute to an inclusive and sustainable ecosystem of existing and new tools for Plasmodium genomic data analysis. From less to more involvement, there are several ways to get involved and join us in contributing to these aims. All contributors must adhere to our community rules and will be recognized as a contributor.
Below are some ways to get involved with current workstreams, but we also have some ideas and plans in the works for the future beyond what is currently available in PGEforge! If you want to contribute in another way that is not currently listed, please reach out to us directly.
Add tool to landscaping and software standards matrices
Landscaping, documenting and benchmarking available Plasmodium genomic data analysis tools and any tools within the realm of Plasmodium genomic epidemiology (PGE) will require continuous updating and curation. As new tools become available, they need to be assessed and integrated into our existing framework. While we have made significant progress in identifying, documenting and evaluating tools commonly applied to Plasmodium genetic data, the tool landscaping and evaluation against software standards is not exhaustive. For example, our initial efforts focused on tools for downstream Plasmodium genetic analysis, specifically targeting tools that extract signals from already processed data and focusing only on tools applicable P. falciparum and P. vivax. In terms of tool evaluation, we focused our efforts on evaluating the tools for which resources were developed in the tutorials section. Future work could also involve other types of tools, however.
So far, PGEforge hosts a landscaping matrix of 40 identified tools, full documentation and evaluation of 17 tools, and complete resources (summary documents, installation instructions, and fully-worked through tutorials) developed for 11 tools.
In order to ensure this ‘live document’ remains up-to-date, we encourage contributions to the following areas:
- Updating the documentation and filling in the gaps for remaining tools in the landscaping matrix
- Evaluating remaining tools based on software standards (these tools are highlighted in grey in the overview by tools based on software standards)
- Updating tool scores against software standards if developers release tool updates
- Add a new tool to landscaping and software standard matrices
To contribute to any of these areas, there are two ways to do this depending on your comfort level with using GitHub and R.
Don’t forget to also add yourself as a contributor!
- Download the relevant
.csvfiles and edit them locally. Then open an issue on Github and attach your updated.csvfiles. The location of the files are described below:
MMS_software_landscaping.csv: this is the tool landscaping matrix.Tools_to_standards.csv: this is the evaluation of each tool with respect to the objective software standards. Please make sure you follow the evaluation criteria rubric when you score each of the criteria.
Please ensure you only modify the relevant cells or add new rows as needed
- Follow the Github contributor guidelines to create your own branch and PR your changes directly. Make sure you:
- Add your headshot photo to the
website_docs/img/peoplefolder - Add your name in alphabetical order to the website_docs/contributors.qmd and reference your photo by using the following code
{fig-align="left" width="100px"}.
Develop resources for a tool
If you have developed a new Plasmodium genomic data analysis tool or identified a tool that is missing tutorial resources, you can develop them and contribute them to PGEforge! Follow these steps to contribute:
Don’t forget to also add yourself as a contributor!
Step 1: In PGEforge we focus on Plasmodium genomic data analysis tools. The tool should be within the scope of Plasmodium genomic epidemiology data analysis.
Step 2: Take a look at the tool landscaping in case we have already evaluated that specific tool. In some instances, we opted to not develop tutorial resources for tools where there was difficulty installing and running the software, run-time errors, or if the tool was clearly superseded by more recent tools. You will find more details in the tool landscaping. If you identify a tool that is not in the landscaping matrix, please add it.
Step 3: If you have reached this point and have found a tool that is missing from PGEforge, great! After adding it to the landscaping and software standards matrices you can develop the tool resources. We provide all the templates that you need to get started. For every tool, we develop the following resources:
- Summary Document: Provide an overview of the tool, including its main purpose, use cases, license, code repository, relevant publications, and citation details. Fill in these details in the template .csv file and then “Render” the accompanying .qmd file to display the information in a nicely formatted table.
- Installation Instructions: Write clear, step-by-step instructions for installing the tool, ensuring it is accessible to users with varying technical skills. If the tool is an R package and not already in the PlasmoGenEpi R-universe, please let us know! Otherwise, we strongly suggest you instruct users to install from R-universe.
- Fully Worked Tutorials: Develop comprehensive tutorials that guide users through the entire process of using the tool to perform a specific analysis, from data import to interpretation of results. PGEforge hosts both empirical and simulated canonical datasets for the commonly used data input formats. We strongly encourage you to use these when you develop these tutorials as this ensures that they are fully reproducible.
You can find the templates for all of these documents in our Github repository and example tutorials in the Tutorials section.
Please make sure to follow our GitHub contribution guidelines.
Add yourself as a contributor
Everyone who contributes to PGEforge is listed as a contributor.
There are two ways to do this:
- Open an issue on Github and make sure you include the following information so we can add you to the contributors list:
- Full name
- Affiliations
- Headshot photo
- Follow the Github contributor guidelines to create your own branch and PR your changes directly. Make sure you:
- Add your headshot photo to the
website_docs/img/peoplefolder - Add your name in alphabetical order to the website_docs/contributors.qmd and reference your photo by using the following code
{fig-align="left" width="100px"}.
Contributing guidelines for GitHub
To ensure a smooth and efficient collaboration process, please follow these guidelines when contributing to our GitHub repository:
- Create a New Branch: Start by creating a new branch from the
developbranch. This keeps your changes separate until they are ready to be reviewed and merged. - Pull Request (PR) for Review: Once you have made your changes, create a PR into the
developbranch. Our team will review your PR and provide feedback or approve it for merging. Never make any changes to themainbranch, and please always PR intodevelop.
Thank you!
Thank you for your interest in contributing to PGEforge. Your efforts help us build a stronger, more inclusive research community making Plasmodium genomic analysis accessible to all!
Community rules
PGEforge is dedicated to creating an inclusive, respectful, and engaging community. We believe in open collaboration and active participation, empowering each other to share and gain knowledge, resources, and opportunities as a community. We believe in the benefits of a wide range of perspectives, experiences and ideas. We welcome contributions from everyone who shares our goals and wants to contribute, regardless of age, gender identity, sexual orientation, disability, ethnicity, nationality, race, religion, education, level of experience, career stage, or socioeconomic status. PGEforge aims to foster a harassment-free experience for everyone and expects all contributors to demonstrate empathy, kindness, and respect, and to engage constructively with differing viewpoints. Unacceptable behaviors, including harassment, discriminatory language, and personal attacks, will not be tolerated. By fostering a positive and inclusive community, we aim to empower everyone to contribute and collaborate effectively to a vibrant and growing PGEforge community.
