Data
Overview of data formats in Plasmodium genomics
Understanding the different types of genomic data formats is essential for anyone involved in Plasmodium genomic analysis, especially beginners or end-users of various analysis tools. Here we briefly cover the primary data formats used in malaria research and common input file formats such as .vcf, .fasta, and others.
Genomic data generation
Before we jump into the different data formats, it is important to understand how malaria genomic data are generated. The variety in data formats arises from different molecular marker panels and available methodologies. The figure below illustrates how different genotyping approaches capture distinct aspects of the Plasmodium genome and how genomic data are ‘generated’ using different techniques and genotyping platforms. Next-generation sequencing is now the most common approach due to significant decreases in cost and its high-throughput nature both for whole genome sequencing but also sequencing of specific molecular markers or genomic regions of interest, also known as targeted sequencing.
Targeted sequencing is similar to WGS, but sample preparation workflow requires an extra step: target enrichment either by hybridization capture or PCR amplicons. These methods are often used for enriching longer and shorter genomic regions, respectively.
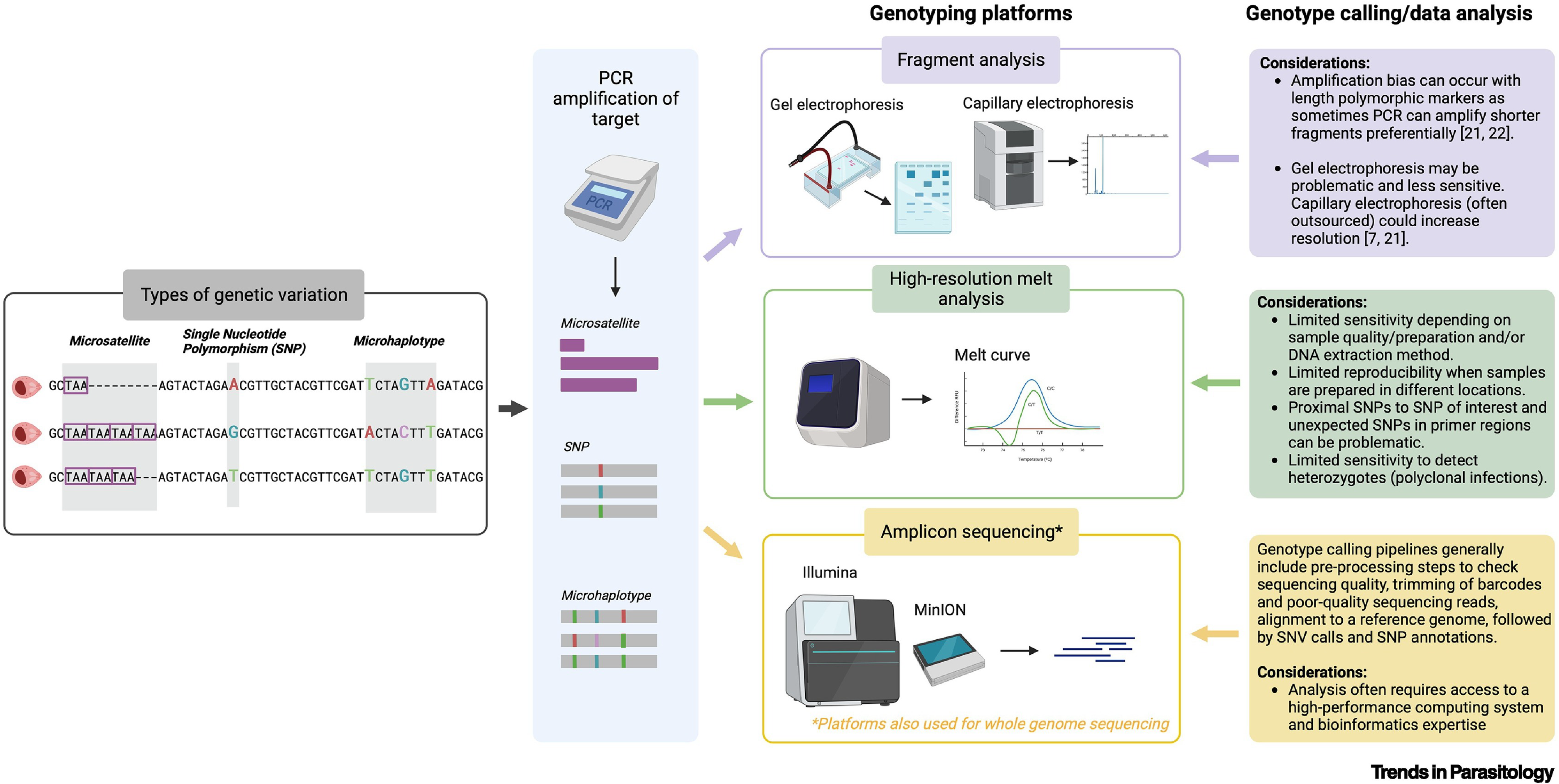
Raw sequencing data is processed by bioinformatic pipelines to call alleles and haplotypes. The outputs of this process, such as VCF, FASTA, or haplotype tables, serve as input formats for downstream data analysis tools.
Data processing
Raw sequence data from next-generation sequencing platforms is processed through bioinformatic pipelines into structured formats suitable for further downstream genomic analysis. Typically this process involves a series of steps. Initially, the raw data undergoes quality control to remove low-quality reads and contaminants. The cleaned data is then aligned to a reference genome, which helps identify the position of each sequence read. From this alignment, variants can be ‘called’, which means variants are identified through differences between the sample and the reference genome. These variants are compiled into Variant Call Format (.vcf) files. Alternatively, the entire cleaned and aligned sequences can be compiled into FASTA format (.fasta) files, which represent the sequences in a text-based format. Additionally, haplotypes may also be called, which means combinations of alleles at multiple loci are identified. These haplotypes are often tabulated in .csv or text-based formats.
Bioinformatic pipelines in malaria genomics need to account for the unique characteristics of Plasmodium genomes, such as high AT content and extensive polymorphism.
Common input formats
When working with Plasmodium genomic data, several input file formats are commonly used across different analysis tools:
.vcf(Variant Call Format): Widely used for storing gene sequence variants, such as SNPs, insertions, deletions, and other types of genetic variants. The standard format includes a mandatory header starting with # or ## in the case of ‘special header keywords’, which provides the metadata such as file format, reference genome. This is followed by the ‘body’, which is tab-separated and each line represents a variant and its relevant information, such as chromosome and position, reference and alternate allele(s), and sample genotype information.
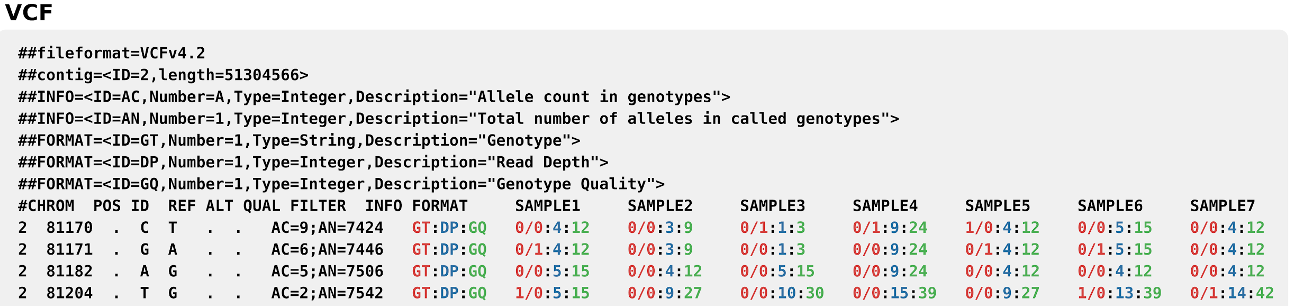
Figure sourced and modified from Wikipedia
.fasta: Widely used for representing nucleotide and protein sequences, in which nucleotides or amino acids are represented using single-letter codes. Each entry begins with a header line starting with > followed by a unique sequence ID and description line, and then the sequence data itself, with one letter per nucleic or amino acid.

Figure sourced from Wikipedia
.txtor.csv: Simple text and comma-separated values formats that are often used to represent haplotype or count data.
Available datasets in PGEforge
As part of the PGEforge community resource, we have compiled simulated and empirical datasets of these common data formats. These datasets are used in the tool tutorials to make them fully reproducible and are freely available for anyone to use.
More details on the datasets hosted on PGEforge can be found in the links below:
They can be accessed at the PGEforge/data folder on Github.
