Microhaplotype data
Microhaplotype data
Mozambique Field Samples
Targeted amplicon data from analysis for the following paper “Sensitive, Highly Multiplexed Sequencing of Microhaplotypes From the Plasmodium falciparum Heterozygome”(Tessema et al. 2022)
This contains 82 field samples gathered from northern and southern Mozambique and had 100 targets (91 diversity targets and 9 targeted drug targets).
The results file can be found within directory amplicon/moz2018_heome1_results_fieldSamples.tsv.gz along with metadata amplicon/moz2018_fieldSamples_meta.tsv. Results are in a 4 column format.
- sample - The name of the sample
- target - The name of the amplicon target
- target_popUID - A population identifier for the haplotype for this target for this sample
- readCnt - The read count for this haplotype for this sample for this target
Lab Control mixtures
Targeted amplicon data from the same 100 target panel as above. Mixtures are made of various combinations of 7 lab strains of P. falciparum and with some mixtures done in replicate at different 4 different parasite densities (10, 100, 1k, 10K.
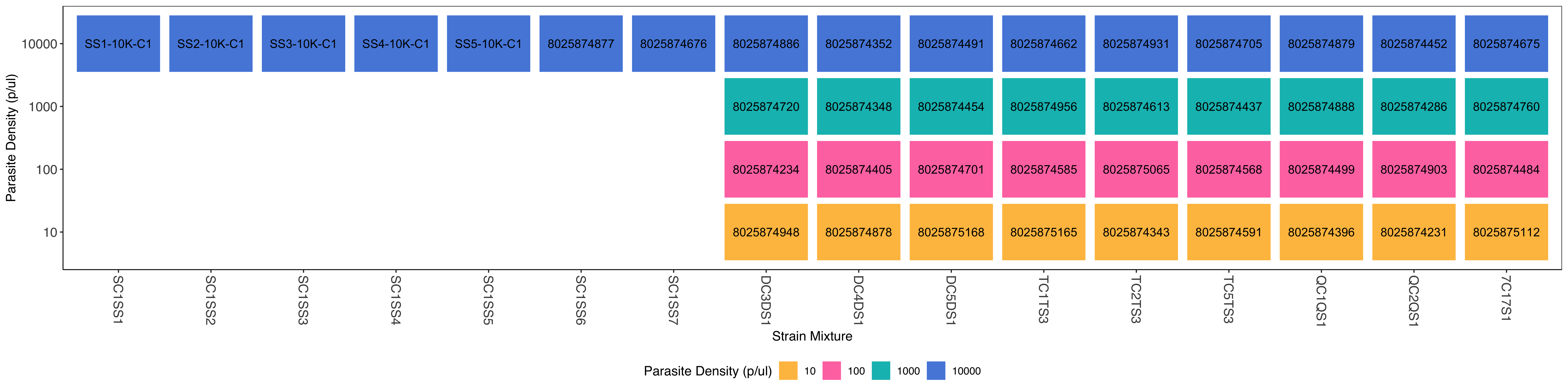
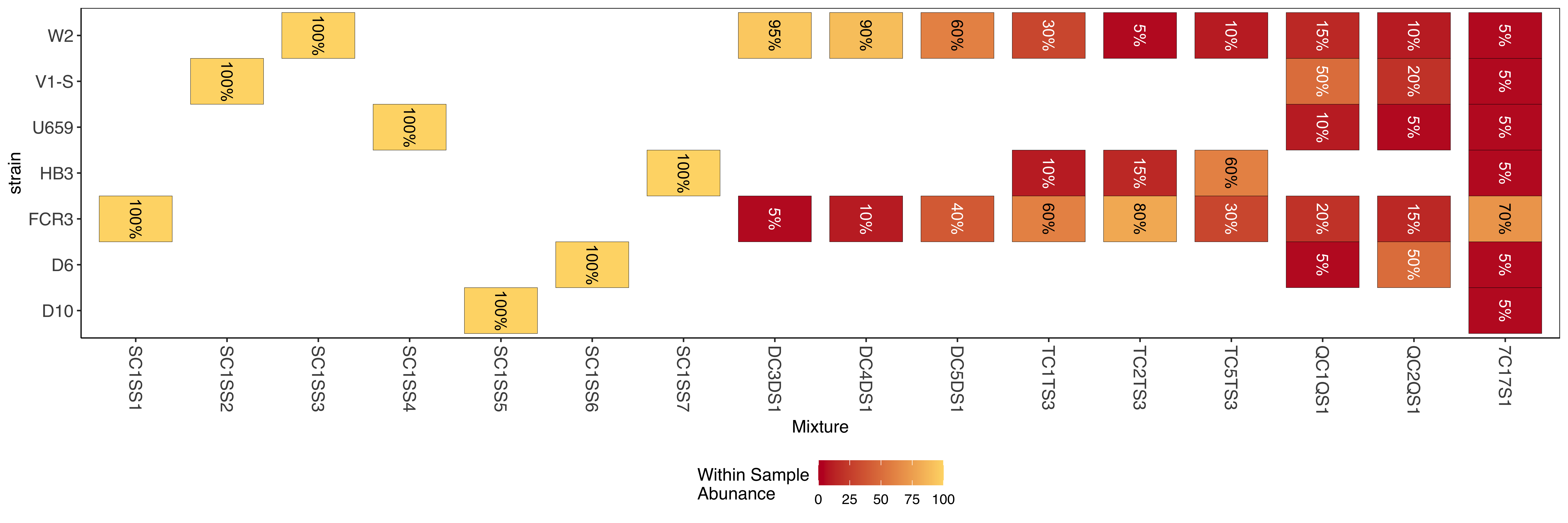
Results are organized in a similar 4 column table as above. The results file can be found within directory amplicon/moz2018_heome1_results_controlSamples.tsv.gz along with metadata amplicon/moz2018_controlSamples_meta.tsv, amplicon/samplesToMixFnp.tab.txt, amplicon/mixSetUpFnp.tab.txt.
Simulated data
Targeted amplicon data was also simulated in silico to create 100 samples sampled from Mozambique and for a newer diversity panel called MAD^4HatTeR with 50 targets selected for thier diversity.
Results are organized in a similar 4 column table as above. The results file can be found within directory amplicon/mozSim_MAD4HATTERDiversitySubPanel.tab.txt.gz
