What’s in store for the future?
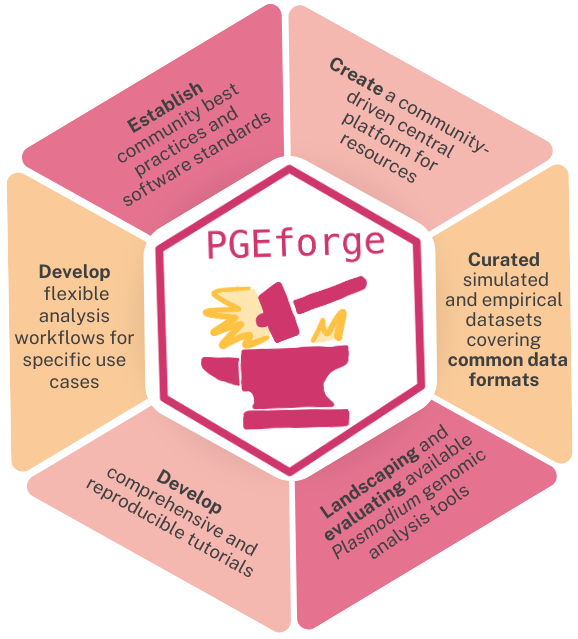
The PGEforge vision
PGEforge is a community-driven platform created by and for malaria genomic epidemiology analysis tool developers and end-users. If you are interested in contributing to these efforts, please get involved!
Tool benchmarking
The process of evaluating tools involves assessing their adherence to objective software standards from both the end-user and developer perspectives. This is a great way to determine how well we are doing at making our tools accessible and usable by anyone. However, it is equally important to evaluate how well tools perform their intended functions and how they compare to other tools designed for the same purpose.
This type of evaluation could be achieved through formal benchmarking of different tools and their analysis functionalities. We can use canonical simulated datasets with known ground truth to evaluate the accuracy, sensitivity, and specificity of the tools in producing desired outcomes. What do we mean by ground truth? For example, we can simulate data to have specific data features that we are interested in benchmarking, including complexity of infection (COI), missingness, sequencing errors and parasitemia. We can then evaluate how well the tools estimate these features. With empirical “real-world” datasets, a ground truth may not be known, but we can use these datasets to compare consistency of each tool obtaining the same outcomes.
By systematically benchmarking tools against known datasets, we can ensure that the tools not only meet software standards but also perform reliably in practical applications, such as key use cases for Plasmodium genomic epidemiology. We plan to host these types of benchmarking resources and results in PGEforge in the near future.
Ongoing work in this space includes benchmarking COI estimation tools.
Adding and evaluating more tools
Our initial efforts to landscape available tools were focused on tools for downstream Plasmodium genetic analysis and included 40 available tools. However, future work could include the following areas (not an exhaustive list!):
- Tools with applications to mosquito genetics, as long as they are generally within the scope of PGEforge (i.e. Plasmodium genomic epidemiology tools)
- Bioinformatic pre-processing tools/pipelines, such as variant calling and quality filters
- Tools that are not specifically engineered for Plasmodium but can be applied to Plasmodiumgenetic data, such as more generic population genetics tools that estimate metrics such as F-statistics, extended haplotype homozygosity, etc
Thank you!
Thank you for your interest in contributing to PGEforge. Your efforts help us build a stronger, more inclusive research community making Plasmodium genomic analysis accessible to all!
