Create a particle_filter object for running
and interacting with a particle filter. A higher-level
interface will be implemented later.
Public fields
modelThe dust model generator being simulated (cannot be re-bound)
n_particlesNumber of particles used (read only)
has_multiple_parametersLogical, indicating if the particle filter requires multiple parameter sets in a list as inputs, and if it it will produce a vector of likelihoods the same length (read only). The parameter sets may or may not use the same data (see
has_multiple_data).has_multiple_dataLogical, indicating if the particle filter simultaneously calculates the likelihood for multiple parameter sets (read only). If
TRUE,has_multiple_parameterswill always beTRUE.n_parametersThe number of parameter sets used by this particle filter (read only). The returned vector of likelihood will be this length, and if
has_multiple_parametersisFALSEthis will be 1.n_dataThe number of data sets used by this particle filter (read only). This will either be 1 or the same value as
n_parameters.
Methods
Method new()
Create the particle filter
Usage
particle_filter$new(
data,
model,
n_particles,
compare,
index = NULL,
initial = NULL,
constant_log_likelihood = NULL,
n_threads = 1L,
seed = NULL,
n_parameters = NULL,
gpu_config = NULL,
stochastic_schedule = NULL,
ode_control = NULL
)Arguments
dataThe data set to be used for the particle filter, created by
particle_filter_data(). This is essentially adata.frame()with at least columnstime_startandtime_end, along with any additional data used in thecomparefunction, and additional information about how your dust time steps relate to a more interpretable measure of model time.modelA stochastic model to use. Must be a
dust_generatorobject.n_particlesThe number of particles to simulate
compareA comparison function. Must take arguments
state,observedandparsas arguments (though the arguments may have different names).stateis the simulated model state (a matrix with as many rows as there are state variables and as many columns as there are particles,datais alistof observed data corresponding to the current time's row in thedataobject provided here in the constructor.parsis any additional parameters passed through to the comparison function (via theparsargument to$run). Alternatively,comparecan beNULLif your model provides a built-in compile compare function (ifmodel$public_methods$has_compare()isTRUE), which may be faster.indexAn index function. This is used to compute the "interesting" indexes of your model. It must be a function of one argument, which will be the result of calling the
$info()method on your model. It should return a list with elementsrun(indices to return at the end of each run, passed through to your compare function) andstate(indices to return if saving state). These indices can overlap but do not have to. This argument is optional but using it will likely speed up your simulation if you have more than a few states as it will reduce the amount of memory copied back and forth.initialA function to generate initial conditions. If given, then this function must accept 3 arguments:
info(the result of calling$info()as forindex),n_particles(the number of particles that the particle filter is using) andpars(parameters passed in in the$runmethod via theparsargument). It must return a list, which can have the elementsstate(initial model state, passed to the particle filter - either a vector or a matrix, and overriding the initial conditions provided by your model) andtime(the initial time step, overriding the first time step of your data - this must occur within your first epoch in yourdataprovided to the constructor, i.e., not less than the first element oftime_startand not more thantime_end). Your function can also return a vector or matrix ofstateand not alter the starting time step, which is equivalent to returninglist(state = state, time = NULL).constant_log_likelihoodAn optional function, taking the model parameters, that computes the constant part of the log-likelihood value (if any). You can use this where your likelihood depends both on the time series (via
data) but also on some non-temporal data. You should bind any non-parameter dependencies into this closure. This is applied at the beginning of the filter run, so represents the initial condition of the marginal log likelihood value propagated by the filter.n_threadsNumber of threads to use when running the simulation. Defaults to 1, and should not be set higher than the number of cores available to the machine.
seedSeed for the random number generator on initial creation. Can be
NULL(to initialise using R's random number generator), a positive integer, or a raw vector - seedust::dustanddust::dust_rngfor more details. Note that the random number stream is unrelated from R's random number generator, except for initialisation withseed = NULL.n_parametersNumber of parameter sets required. This, along with
data, controls the interpretation of how the particle filter, and importantly will add an additional dimension to most outputs (scalars become vectors, vectors become matrices etc).gpu_configGPU configuration, typically an integer indicating the device to use, where the model has GPU support. An error is thrown if the device id given is larger than those reported to be available (note that CUDA numbers devices from 0, so that '0' is the first device, so on). See the method
$gpu_info()for available device ids; this can be called before object creation asmodel$public_methods$gpu_info(). For additional control, provide a list with elementsdevice_idandrun_block_size. Further options (and validation) of this list will be added in a future version!stochastic_scheduleVector of times to perform stochastic updates, for continuous time models.
ode_controlTuning control for the ODE stepper, for continuous time (ODE) models
Method run()
Run the particle filter
Usage
particle_filter$run(
pars = list(),
save_history = FALSE,
save_restart = NULL,
min_log_likelihood = NULL
)Arguments
parsA list representing parameters. This will be passed as the
parsargument to your model, to yourcomparefunction, and (if using) to yourinitialfunction. It must be an R list (not vector orNULL) because that is what a dust model currently requires on initialisation or$reset- we may relax this later. You may want to put your observation and initial parameters under their own keys (e.g.,pars$initial$whatever), but this is up to you. Extra keys are silently ignored by dust models.save_historyLogical, indicating if the history of all particles should be saved. If saving history, then it can be queried later with the
$historymethod on the object.save_restartAn integer vector of time points to save restart infomation for. These are in terms of your underlying time variable (the
timecolumn inparticle_filter_data()) not in terms of time steps. The state will be saved after the particle filtering operation (i.e., at the end of the step).min_log_likelihoodOptionally, a numeric value representing the smallest likelihood we are interested in. If given and the particle filter drops below this number, then we terminate early and return
-Inf. In this case, history and final state cannot be returned from the filter. This is primarily intended for use with pmcmc where we can avoid computing likelihoods that will certainly be rejected. Only suitable for use where log-likelihood increments (with thecomparefunction) are always negative. This is the case if you use a normalised discrete distribution, but not necessarily otherwise. If using a multi-parameter filter this can be a single number (in which case the exit is when the sum of log-likelihoods drops below this threshold) or a vector of numbers the same length aspars(in which case exit occurs when all numbers drop below this threshold).
Method run_begin()
Begin a particle filter run. This is part of the
"advanced" interface for the particle filter; typically you will
want to use $run() which provides a user-facing wrapper around
this function. Once created with $run_begin(), you should take
as many steps as needed with $step().
Usage
particle_filter$run_begin(
pars = list(),
save_history = FALSE,
save_restart = NULL,
min_log_likelihood = NULL
)Arguments
parsA list representing parameters. See
$run()for details.save_historyLogical, indicating if the history of all particles should be saved. See
$run()for details.save_restartTimes to save restart state at. See
$run()for details.min_log_likelihoodOptionally, a numeric value representing the smallest likelihood we are interested in. See
$run()for details.
Method state()
Extract the current model state, optionally filtering. If the model has not yet been run, then this method will throw an error. Returns a matrix with the number of rows being the number of model states, and the number of columns being the number of particles.
Method history()
Extract the particle trajectories. Requires that
the model was run with save_history = TRUE, which does
incur a performance cost. This method will throw an error if
the model has not run, or was run without save_history = TRUE. Returns a 3d array with dimensions corresponding to (1)
model state, filtered by index$run if provided, (2)
particle (following index_particle if provided), (3)
time point. If using a multi-parameter filter then returns a 4d array
with dimensions corresponding to (1) model state, (2) particle, (3)
parameter, (4) time point.
Method ode_statistics()
Fetch statistics about steps taken during the integration, by
calling through to the $statistics() method of the underlying
model. This is only available for continuous time (ODE) models,
and will error if used with discrete time models.
Method restart_state()
Return the full particle filter state at points back in time
that were saved with the save_restart argument to
$run(). If available, this will return a 3d array, with
dimensions representing (1) particle state, (2) particle index,
(3) time point. If multiple parameters are used then returns a 4d array,
with dimensions representing (1) particle state, (2) particle index,
(3) parameter set, (4) time point. This could be quite large, especially
if you are using the index argument to create the particle filter
and return a subset of all state generally. It is also
different to the saved trajectories returned by $history()
because earlier saved state is not filtered by later filtering
(in the history we return the tree of history representing the
histories of the final particles, here we are returning all
particles at the requested point, regardless if they appear in
the set of particles that make it to the end of the
simulation).
Method inputs()
Return a list of inputs used to configure the particle
filter. These correspond directly to the argument names for the
particle filter constructor and are the same as the input
argument with the exception of seed, which is the state of
the rng if it has been used (this can be used as a seed to
restart the model).
Method set_n_threads()
Set the number of threads used by the particle filter (and dust model) after creation. This can be used to allocate additional (or subtract excess) computing power from a particle filter. Returns (invisibly) the previous value.
Arguments
n_threadsThe new number of threads to use. You may want to wrap this argument in
dust::dust_openmp_threads()in order to verify that you can actually use the number of threads requested (based on environment variables and OpenMP support).
Examples
# A basic SIR model included in the dust package
gen <- dust::dust_example("sir")
# Some data that we will fit to, using 1 particle:
sir <- gen$new(pars = list(), time = 0, n_particles = 1)
dt <- 1 / 4
day <- seq(1, 100)
incidence <- rep(NA, length(day))
true_history <- array(NA_real_, c(5, 1, 101))
true_history[, 1, 1] <- sir$state()
for (i in day) {
state_start <- sir$state()
sir$run(i / dt)
state_end <- sir$state()
true_history[, 1, i + 1] <- state_end
# Reduction in S
incidence[i] <- state_start[1, 1] - state_end[1, 1]
}
# Convert this into our required format:
data_raw <- data.frame(day = day, incidence = incidence)
data <- particle_filter_data(data_raw, "day", 4, 0)
# A comparison function
compare <- function(state, observed, pars = NULL) {
if (is.null(pars$exp_noise)) {
exp_noise <- 1e6
} else {
exp_noise <- pars$exp_noise
}
incidence_modelled <- state[1,]
incidence_observed <- observed$incidence
lambda <- incidence_modelled +
rexp(length(incidence_modelled), exp_noise)
dpois(incidence_observed, lambda, log = TRUE)
}
# Construct the particle_filter object with 100 particles
p <- particle_filter$new(data, gen, 100, compare)
p$run(save_history = TRUE)
#> [1] -17249.82
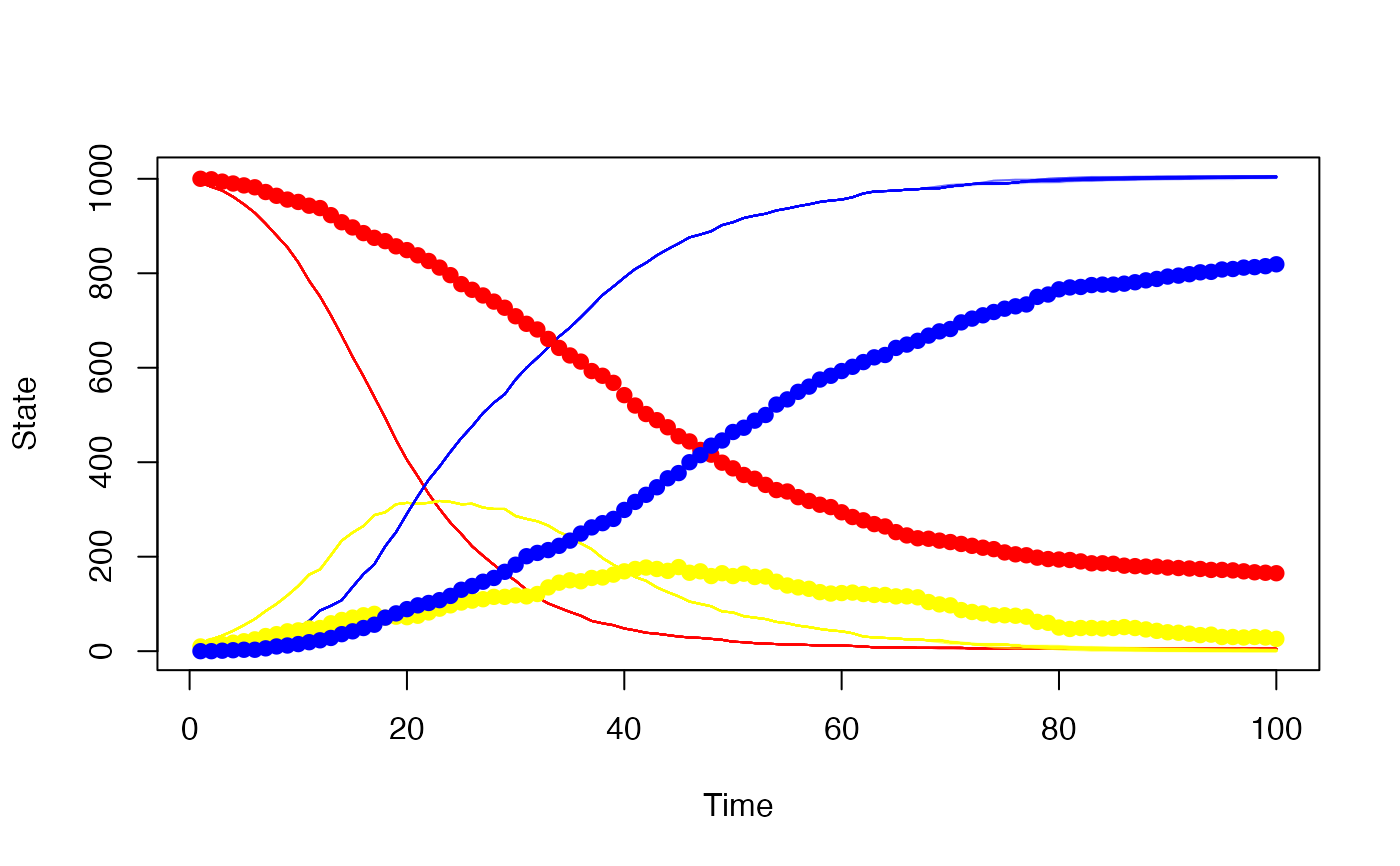
# Our simulated trajectories, with the "real" data superimposed
history <- p$history()
matplot(data_raw$day, t(history[1, , -1]), type = "l",
xlab = "Time", ylab = "State",
col = "#ff000022", lty = 1, ylim = range(history))
matlines(data_raw$day, t(history[2, , -1]), col = "#ffff0022", lty = 1)
matlines(data_raw$day, t(history[3, , -1]), col = "#0000ff22", lty = 1)
matpoints(data_raw$day, t(true_history[1:3, , -1]), pch = 19,
col = c("red", "yellow", "blue"))