Return prior
Bob Verity and Pete Winskill
2024-06-26
Source:vignettes/checks_return_prior.Rmd
checks_return_prior.Rmd## Registered S3 method overwritten by 'GGally':
## method from
## +.gg ggplot2Purpose: to check that drjacoby returns the prior distribution when no likelihood is used.
Model
Four parameters, each representing a different one of the four possible parameter transformations internally. Each parameter is given a suitable informative prior over it’s domain.
Parameters dataframe:
df_params <- define_params(name = "real_line", min = -Inf, max = Inf,
name = "neg_line", min = -Inf, max = 0,
name = "pos_line", min = 0, max = Inf,
name = "unit_interval", min = 0, max = 1)Likelihood and prior:
Plots
output_sub <- subset(mcmc$output, phase == "sampling")
# real_line
hist(output_sub$real_line, breaks = 100, probability = TRUE, col = "black", main = "real_line")
x <- seq(-5, 5, l = 1001)
lines(x, dnorm(x), col = 2, lwd = 2)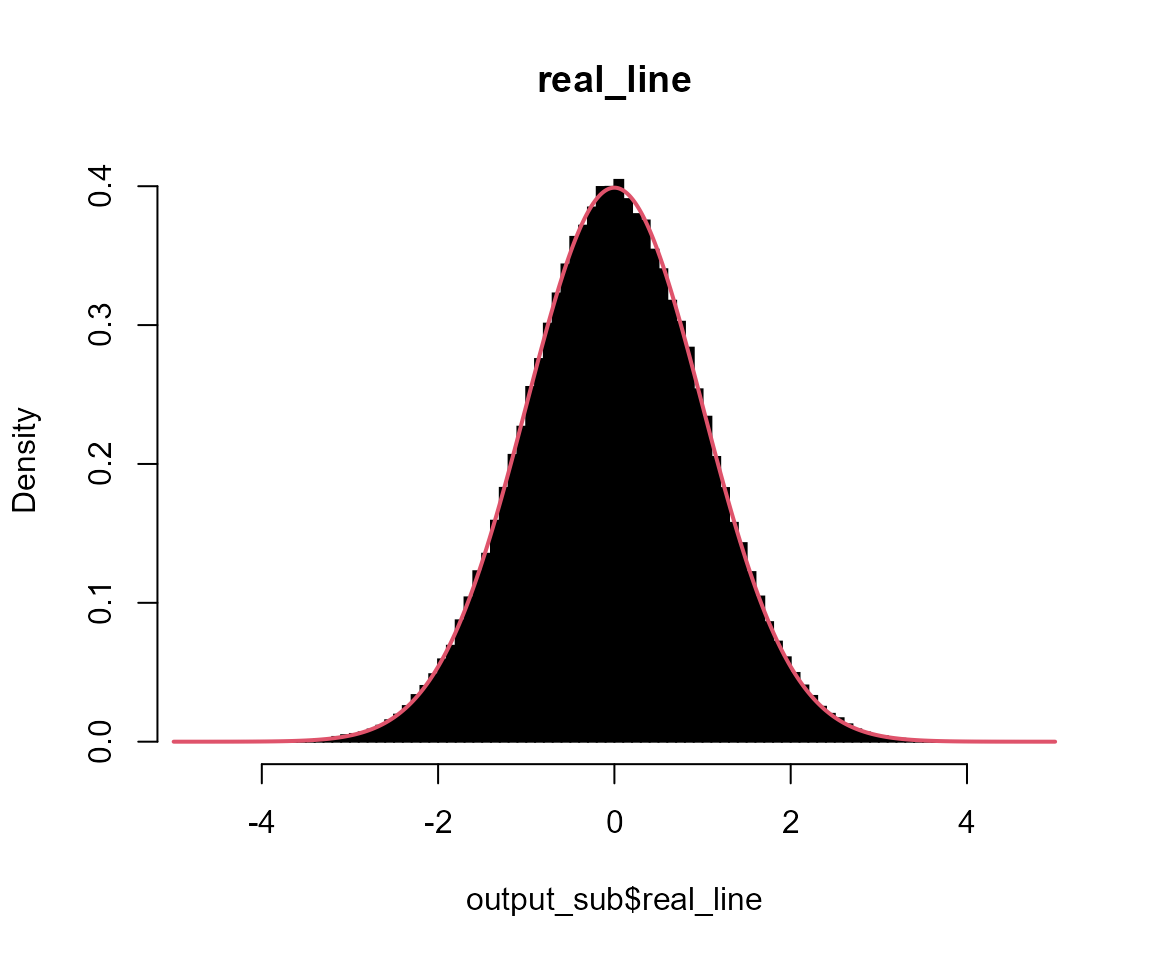
# neg_line
hist(output_sub$neg_line, breaks = 100, probability = TRUE, col = "black", main = "neg_line")
x <- seq(-100, 0, l = 1001)
lines(x, dgamma(-x, shape = 5, scale = 5), col = 2, lwd = 2)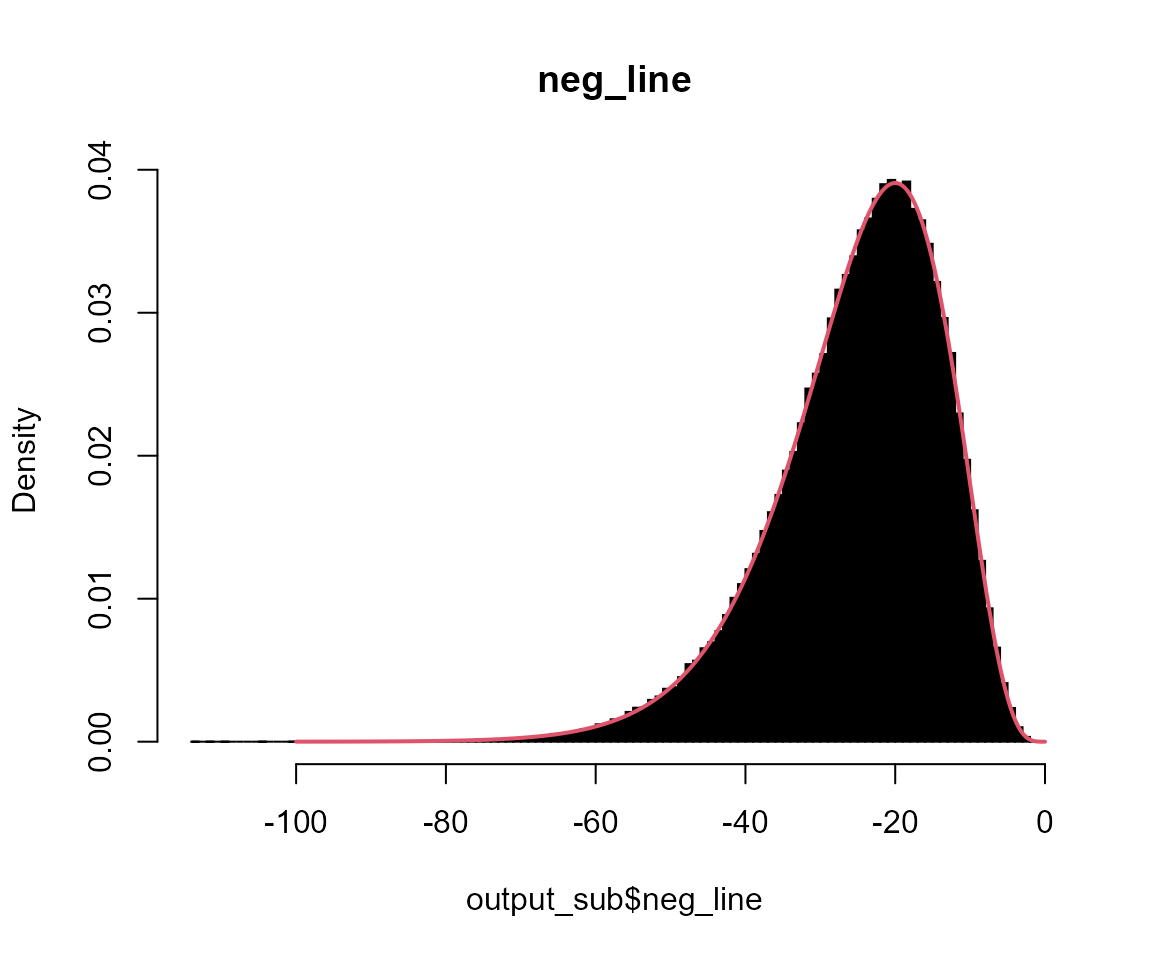
# pos_line
hist(output_sub$pos_line, breaks = 100, probability = TRUE, col = "black", main = "pos_line")
x <- seq(0, 100, l = 1001)
lines(x, dgamma(x, shape = 5, scale = 5), col = 2, lwd = 2)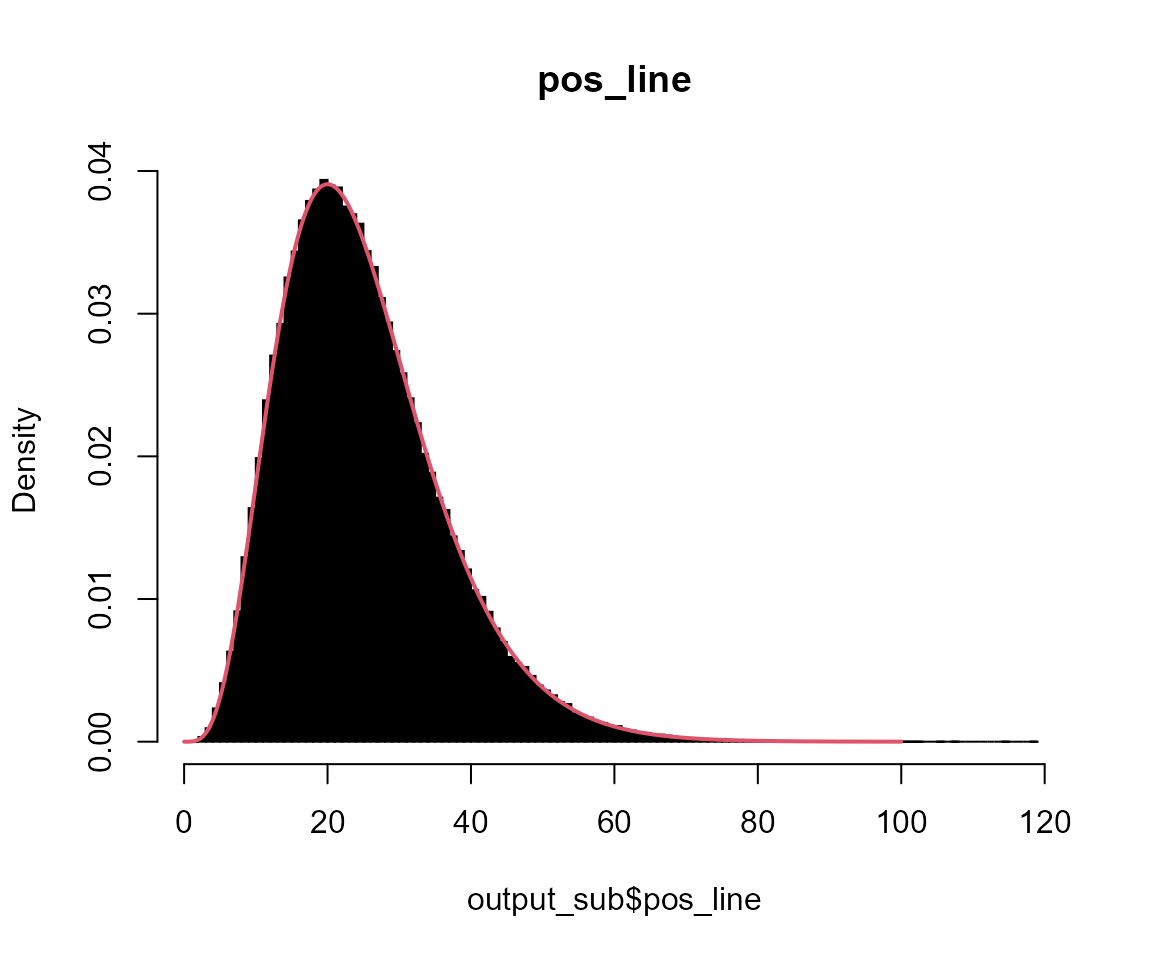
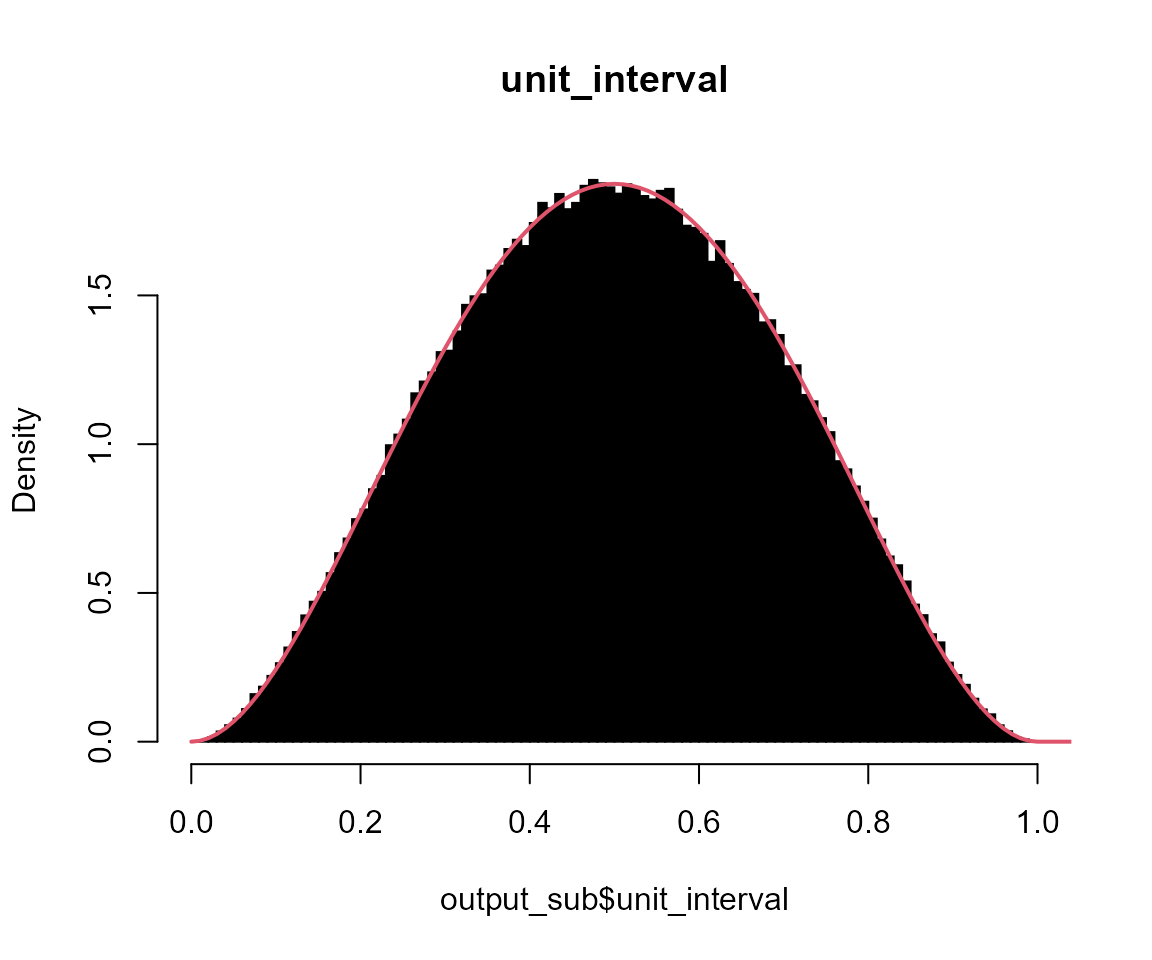
# unit_interval
hist(output_sub$unit_interval, breaks = seq(0, 1, 0.01), probability = TRUE, col = "black", main = "unit_interval")
x <- seq(0, 10, l = 1001)
lines(x, dbeta(x, shape1 = 3, shape2 = 3), col = 2, lwd = 2)