Dr. Jacoby 
drjacoby is a package for running flexible Markov chain Monte Carlo (MCMC) with minimal fiddling required by the user. The likelihood and the priors that go into the model can be written as either R or C++ functions, with the latter typically being much faster to run. Outputs are produced in a standardised format, and can be explored using a range of built in diagnostic plots and statistics.
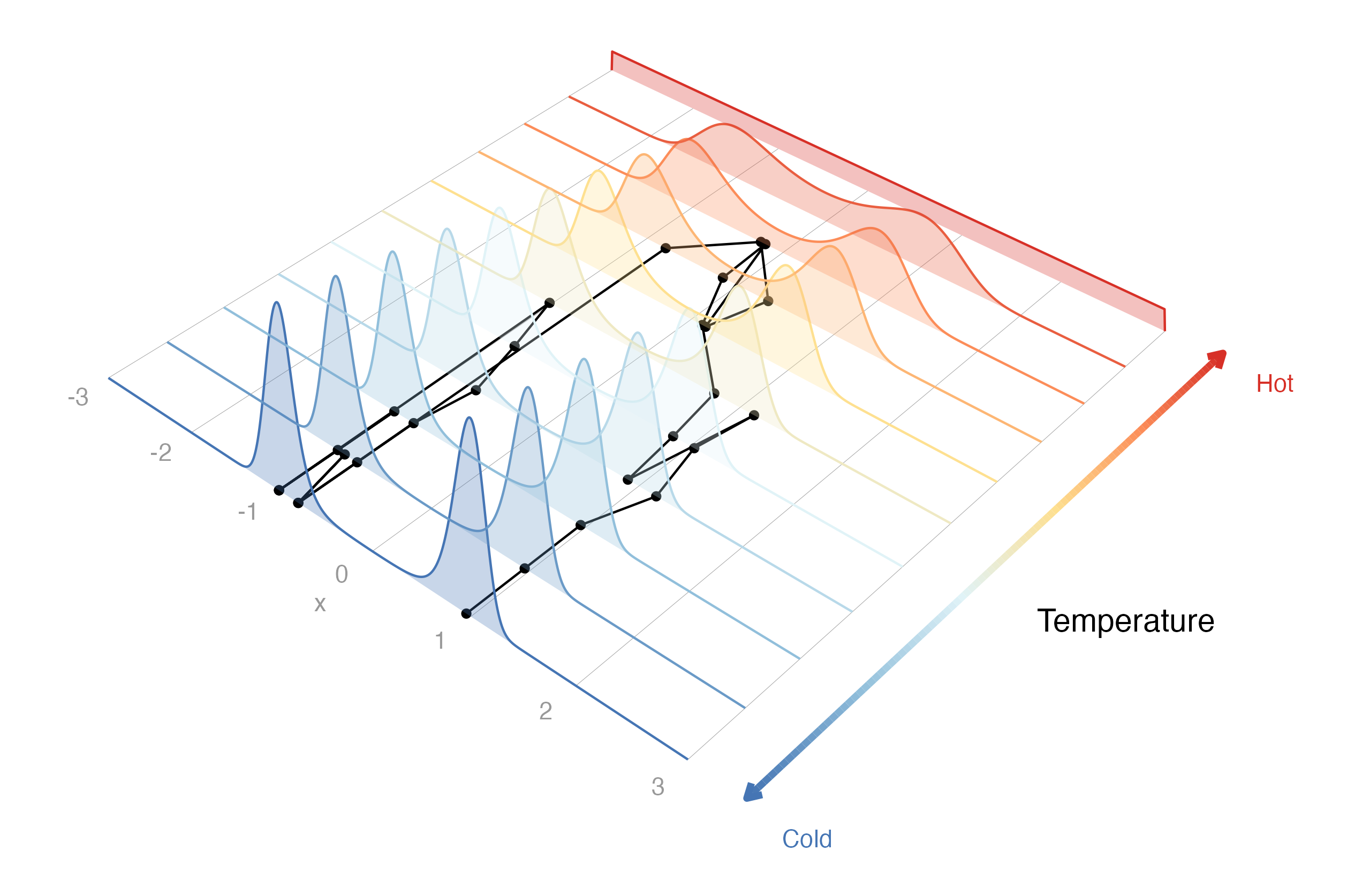
drjacoby uses parallel tempering to “melt” the target distribution,
thereby allowing information to pass between well-separated peaks.
There are many MCMC programs out there, including more far-reaching programs such as WinBUGS, JAGS, greta, STAN, and many more. These programs contain a wide variety of options for specifying complex models, and often run different flavours of MCMC such as Hamiltonian Monte Carlo (HMC). In contrast, drjacoby is tailored to a specific type of MCMC problem: those that mix poorly due to highly correlated and/or multi-modal posteriors, which can occur in both simple and complex models. If the posterior is particularly peaky then even methods like HMC may fail, meaning (unfortunately) no amount of STAN iterations will get us to the right answer. There are techniques available for dealing with this sort of problem, but they tend to be fairly advanced and tricky to implement. The aim of drjacoby is to bring these methods within reach of a non-specialist MCMC user, so that reliable results can be obtained without the need to code a custom MCMC program from scratch.
After installing drjacoby, please take at look at the first example application to get up and running.