Efficacy
Efficacy.RmdWe can parameterise the mode of action of the vaccine to be infection-blocking, anti-disease or both.
Infection-blocking vaccine
Next, we can run with an infection-blocking vaccine:
# Run the determinstic model with an example population and infection-blocking vaccine
infection_blocking <- run(country = "United Kingdom",
max_vaccine = 200000,
vaccine_efficacy_disease = rep(0, 17),
vaccine_efficacy_infection = rep(0.9, 17))
# Format the output selecting infection and deaths
out2 <- format(infection_blocking, compartments = NULL, summaries = c("infections", "deaths")) %>%
mutate(Name = "Infection blocking")Anti-disease vaccine
And finally, a run with a disease-blocking vaccine:
# Run the determinstic model with an example population and anti-disease vaccine
disease_blocking <- run(country = "United Kingdom",
max_vaccine = 200000,
vaccine_efficacy_disease = rep(0.9, 17),
vaccine_efficacy_infection = rep(0, 17))
# Format the output selecting infection and deaths
out3 <- format(disease_blocking, compartments = NULL, summaries = c("infections", "deaths")) %>%
mutate(Name = "Disease blocking")Compare
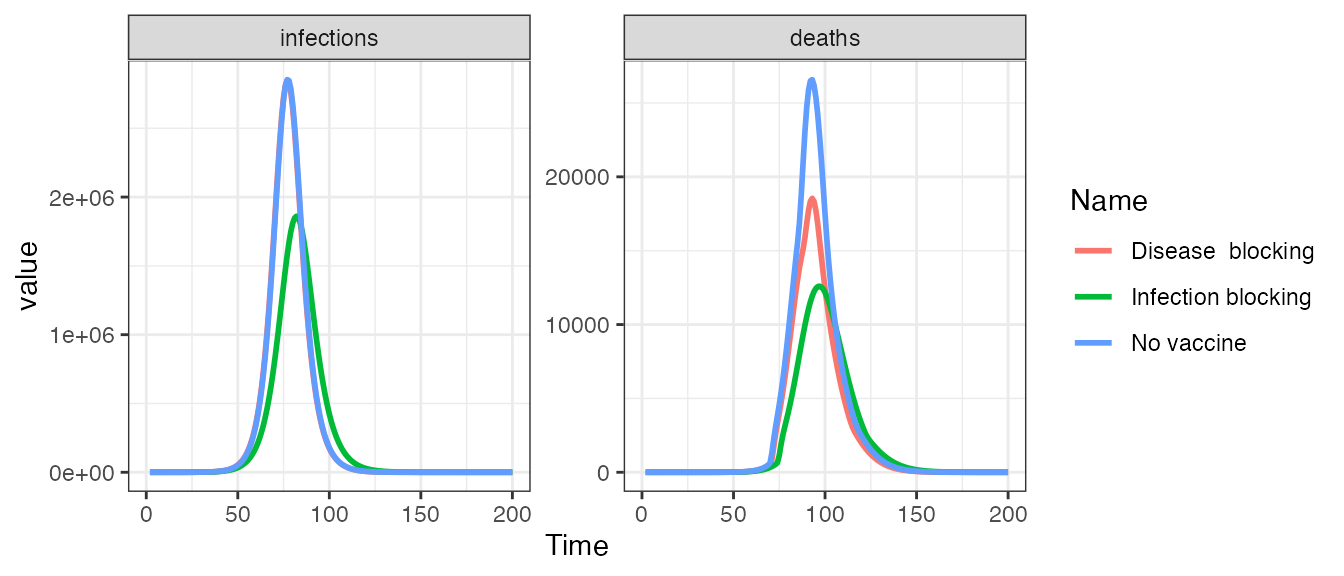
# Create plot data.frame
pd <- bind_rows(out1, out2, out3)
# Plot outputs
ggplot(pd, aes(x = t, y = value, group = Name, col = Name)) +
geom_line(size = 1) +
facet_wrap(~ compartment, scales = "free_y", ncol = 2) +
xlim(0, 200) +
xlab("Time") +
theme_bw()
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.
#> Warning: Removed 996 rows containing missing values or values outside the scale range
#> (`geom_line()`).