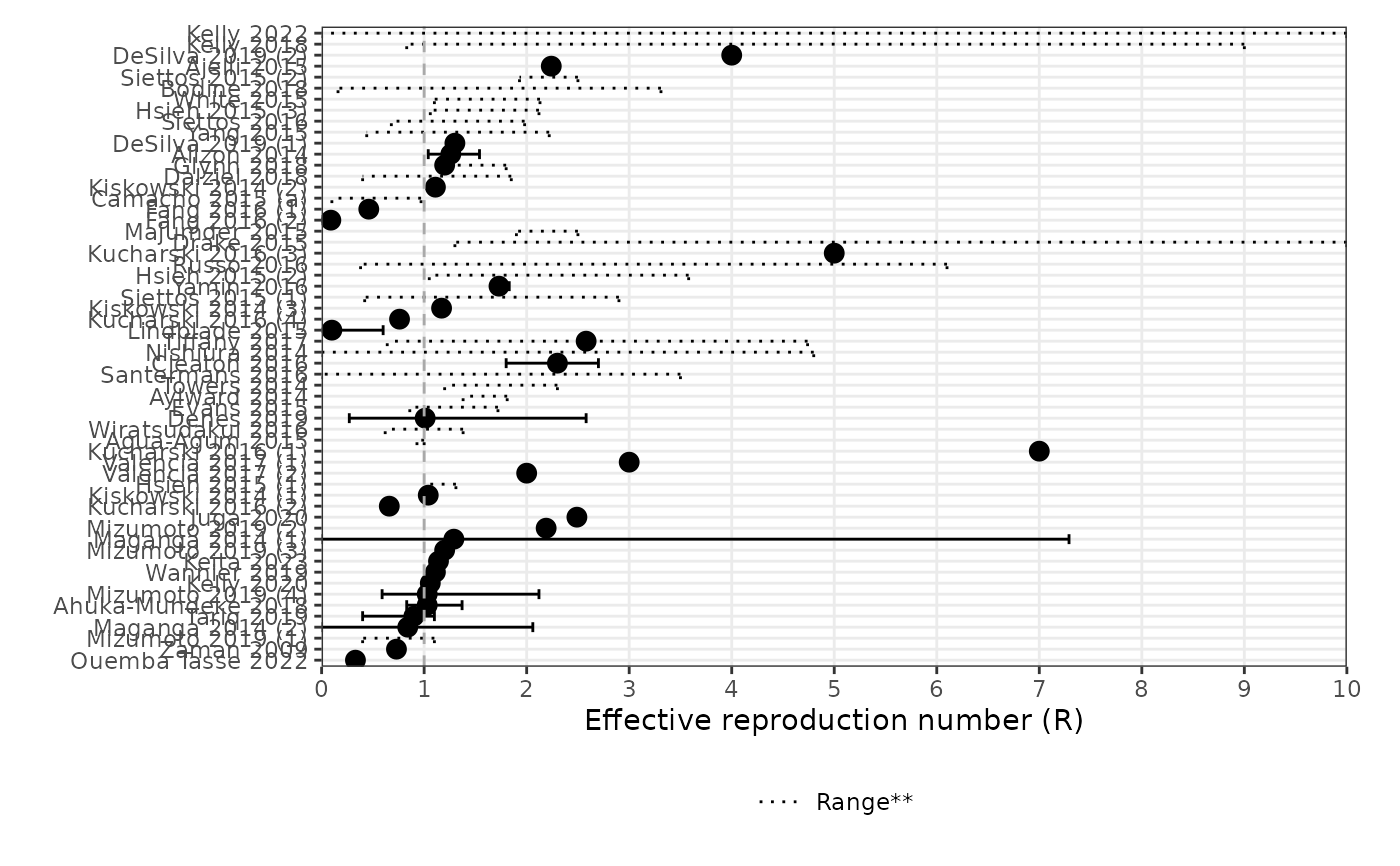
Generate a forest plot for effective reproduction number (Rt)
Source:R/forest_plot_r.R
forest_plot_rt.RdThis function generates a forest plot for the effective reproduction number (Rt) using the provided data frame.
Arguments
- df
The data frame containing the necessary data for generating the forest plot.
- ulim
The upper limit for the x-axis of the plot. Default is 10.
- reorder_studies
Logical. If TRUE, the studies will be reordered using the
reorder_studiesfunction. Default is TRUE.- ...
Additional arguments to be passed to the
forest_plotfunction.
Examples
df <- load_epidata("ebola")[["params"]]
#> ℹ ebola does not have any extracted outbreaks
#> information. Outbreaks will be set to NULL.
#> ✔ Data loaded for ebola
forest_plot_rt(df)
#> Warning: Removed 24 rows containing missing values or values outside the scale range
#> (`geom_point()`).