ebola <- load_epidata('ebola')
#> ℹ ebola does not have any extracted outbreaks
#> information. Outbreaks will be set to NULL.
#> ✔ Data loaded for ebola
params <- ebola[["params"]]
params <- filter_cols(params, "article_qa_score", funs = ">", vals = "50")
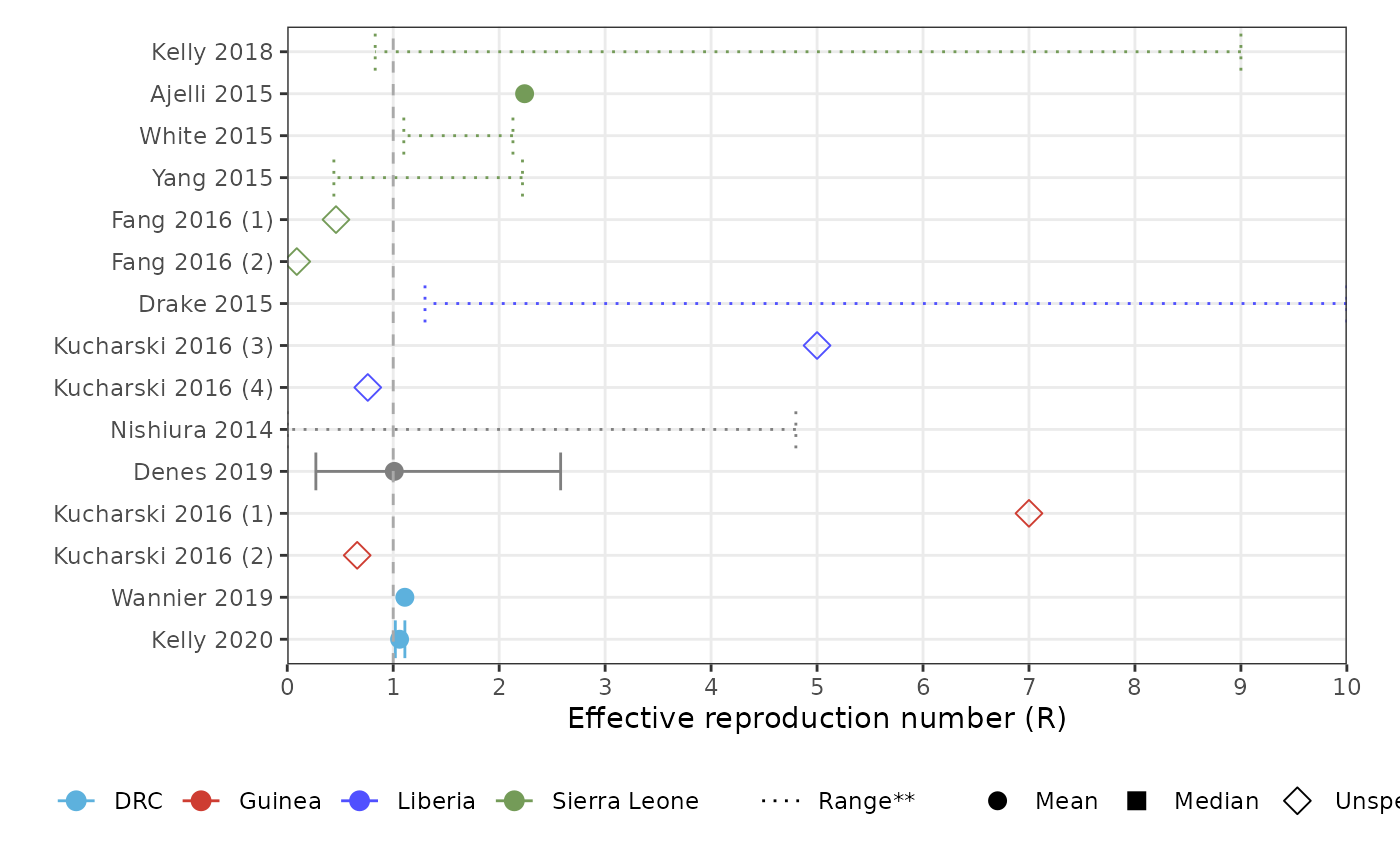
forest_plot_rt(params, col_by = "population_country", shape_by = "parameter_value_type")
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_point()`).
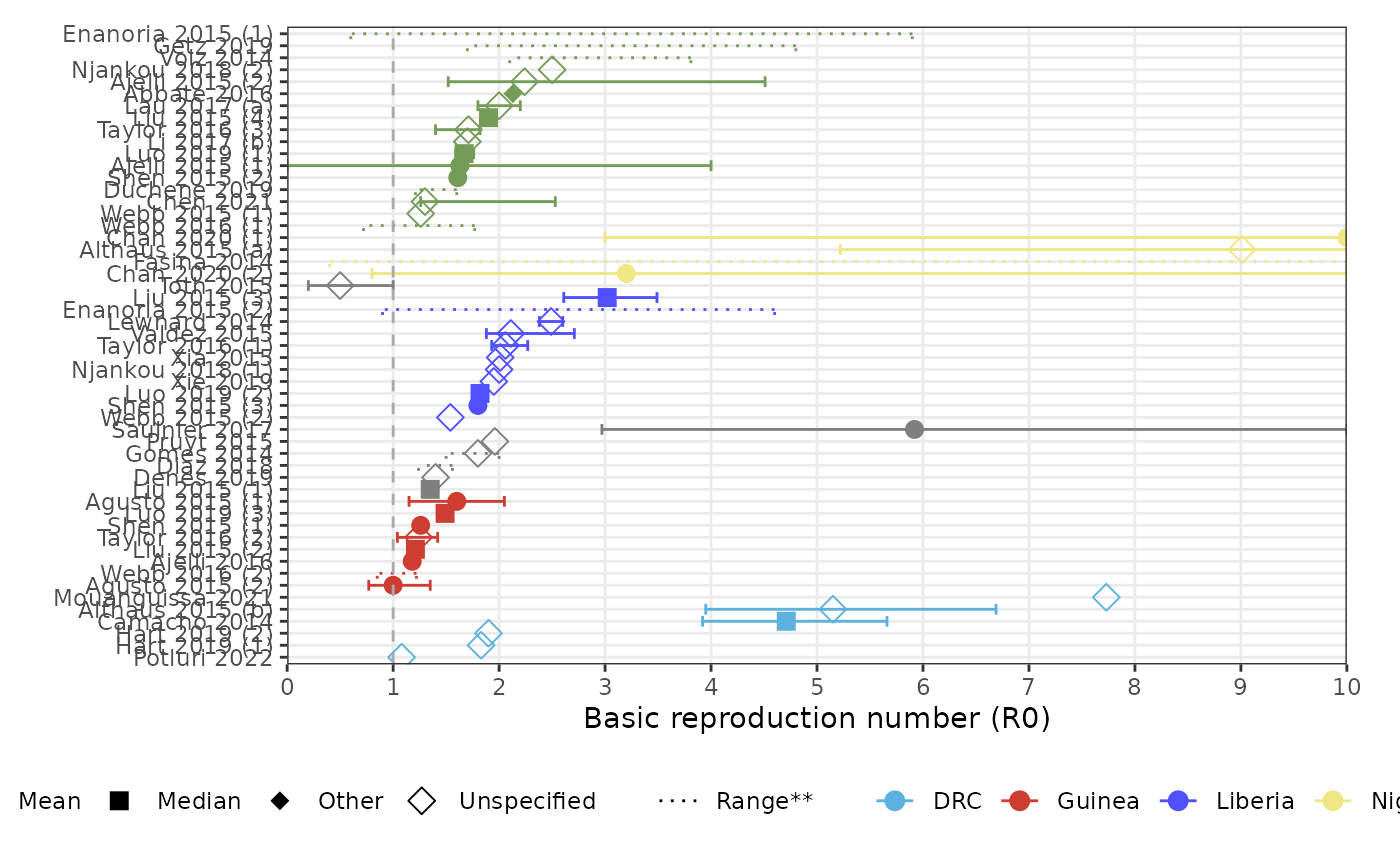
forest_plot_r0(params, col_by = "population_country", shape_by = "parameter_value_type")
#> Warning: Removed 9 rows containing missing values or values outside the scale range
#> (`geom_point()`).