odin2 implements a high-level language for describing
and implementing ordinary differential equations and difference
equations in R. It provides a “domain
specific language” (DSL) which looks like R but is compiled
directly to C++, using dust2 to solve
your system and to provide an interface to particle filters. You can
then use monty to fit
your models using MCMC.
This vignette jumps through a few of the core features of
odin2 and ways that you might use it with
dust2 and monty. The other vignettes and
the odin & monty
book expand on topics covered here in more detail.
Discrete time stochastic SIR model
A simple definition of the SIR model is:
where is the number of susceptibles, is the number of infected and is the number recovered; the total population size is constant. is the infection rate, is the recovery rate.
Discretising this model in time steps of width gives the following update equations for each time step:
where
Here is this system, as a stochastic compartmental model:
gen <- odin2::odin({
p_IR <- 1 - exp(-gamma * dt)
N <- parameter(1000)
p_SI <- 1 - exp(-(beta * I / N * dt))
n_SI <- Binomial(S, p_SI)
n_IR <- Binomial(I, p_IR)
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
initial(S) <- N - I0
initial(I) <- I0
initial(R) <- 0
beta <- parameter(0.2)
gamma <- parameter(0.1)
I0 <- parameter(10)
})
#> ✔ Wrote 'DESCRIPTION'
#> ✔ Wrote 'NAMESPACE'
#> ✔ Wrote 'R/dust.R'
#> ✔ Wrote 'src/dust.cpp'
#> ✔ Wrote 'src/Makevars'
#> ℹ 12 functions decorated with [[cpp11::register]]
#> ✔ generated file cpp11.R
#> ✔ generated file cpp11.cpp
#> ℹ Re-compiling odin.system9a8a577a
#> ── R CMD INSTALL ───────────────────────────────────────────────────────────────
#> * installing *source* package ‘odin.system9a8a577a’ ...
#> ** this is package ‘odin.system9a8a577a’ version ‘0.0.1’
#> ** using staged installation
#> ** libs
#> using C++ compiler: ‘g++ (Ubuntu 13.3.0-6ubuntu2~24.04) 13.3.0’
#> g++ -std=gnu++17 -I"/opt/R/4.5.2/lib/R/include" -DNDEBUG -I'/home/runner/work/_temp/Library/cpp11/include' -I'/home/runner/work/_temp/Library/dust2/include' -I'/home/runner/work/_temp/Library/monty/include' -I/usr/local/include -DHAVE_INLINE -fopenmp -fpic -g -O2 -Wall -pedantic -fdiagnostics-color=always -c cpp11.cpp -o cpp11.o
#> g++ -std=gnu++17 -I"/opt/R/4.5.2/lib/R/include" -DNDEBUG -I'/home/runner/work/_temp/Library/cpp11/include' -I'/home/runner/work/_temp/Library/dust2/include' -I'/home/runner/work/_temp/Library/monty/include' -I/usr/local/include -DHAVE_INLINE -fopenmp -fpic -g -O2 -Wall -pedantic -fdiagnostics-color=always -c dust.cpp -o dust.o
#> g++ -std=gnu++17 -shared -L/opt/R/4.5.2/lib/R/lib -L/usr/local/lib -o odin.system9a8a577a.so cpp11.o dust.o -fopenmp -L/opt/R/4.5.2/lib/R/lib -lR
#> installing to /tmp/RtmpR87IYe/devtools_install_1f8c61fa0e9c/00LOCK-dust_1f8c6f949403/00new/odin.system9a8a577a/libs
#> ** checking absolute paths in shared objects and dynamic libraries
#> * DONE (odin.system9a8a577a)
#> ℹ Loading odin.system9a8a577aThis step generates C++ code for the model and compiles it; it will take a few seconds.
Once we have our system, we can pass it to
[dust2::dust_system_create] to create and start simulating
it. Our system above has defaults for its parameters (N,
beta, gamma, and I0) so we can
initialise with almost no arguments:
pars <- list(beta = 0.2, gamma = 0.1, I0 = 10, N = 1000)
sys <- dust2::dust_system_create(gen(), pars, n_particles = 10)By default the system will start at time 0 and with
dt = 1. We can simulate 10 random epidemics starting from
our initial conditions:
dust2::dust_system_set_state_initial(sys)
time <- 0:100
y <- dust2::dust_system_simulate(sys, time)
matplot(time, t(y[2, , ]), col = "#00000055", lty = 1, type = "l",
xlab = "Time", ylab = "Number of infecteds")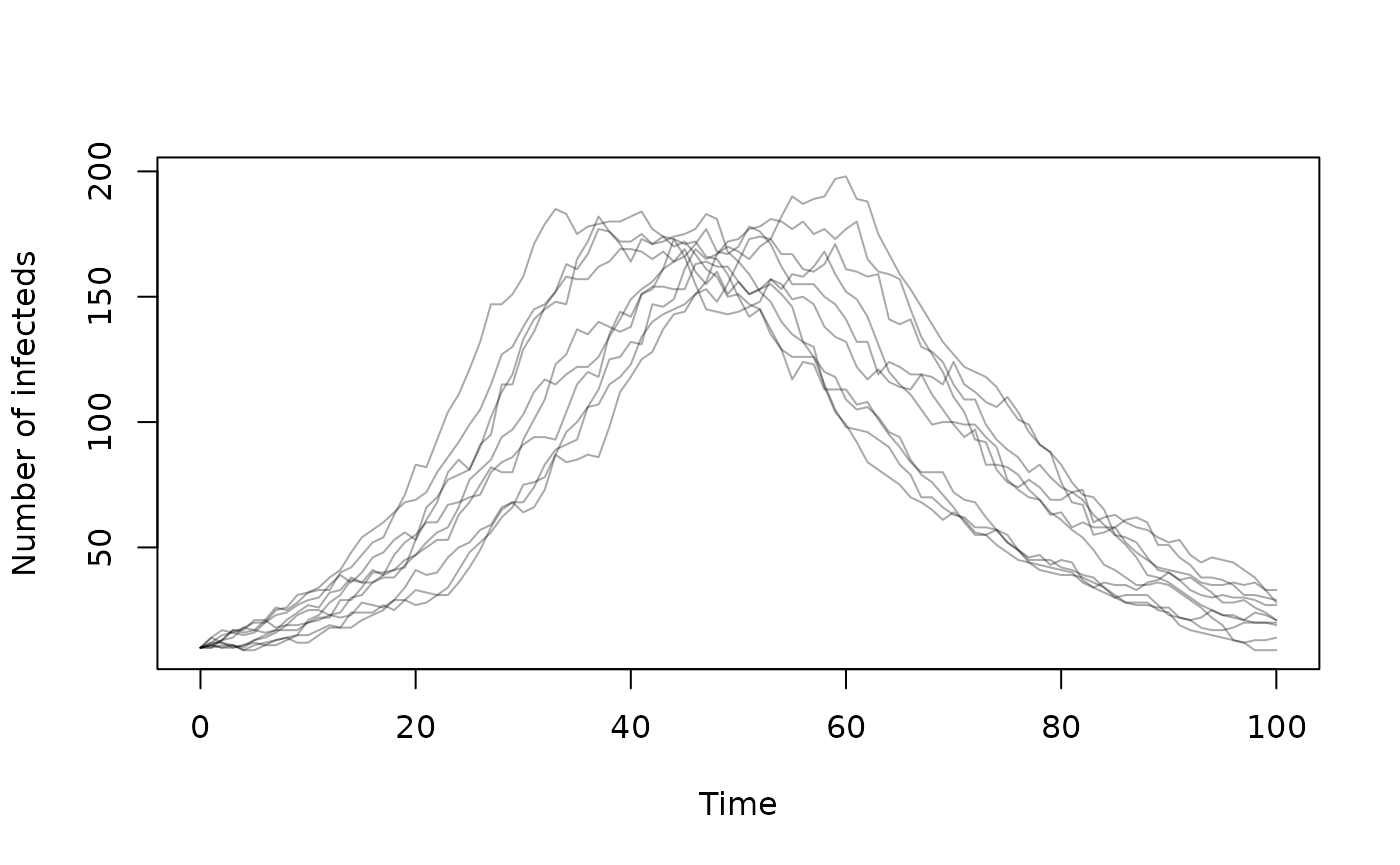
as this system is stochastic, each trajectory will be different.
