Introduction
This package defines a set of useful primitives for structuring and simulating individual based mechanistic models, with special emphasis on infectious disease modelling. Structuring means that the package lets you specify the state space of the model and transition rules for how state changes over time, and simulating means that the package defines a principled way to put state and transitions together to update state over a time step. Transitions are any functions that change state, and in “individual” come in two forms: processes, which occur each time step and events, which are scheduled. The simulation updates over a discrete time step of unit length.
This vignette provides a description of the various primitive
elements provided by the “individual” package for modelling and explains
how they work together. Please see the vignette("Tutorial")
for a practical example.
Variables
In “individual”, variables are how one defines state, and represent any attribute of an individual. While many variables will be dynamically updated throughout a simulation, they don’t have to be. Specifying a baseline characteristic for each individual that doesn’t change over a simulation will still be specified using a variable object.
There are 3 types of variable objects:
individual::CategoricalVariable can represent a set of
mutually exclusive categories, individual::IntegerVariable
for representing integers, and individual::DoubleVariable
for continuous values (real numbers).
Categorical Variable
Most epidemiological models will require use of categorical
variables to define state, for example, the Susceptible, Infectious, and
Recovered classes in the classic SIR
model. In general, for compartmental models, each set of
compartments which describes an aspect of an individual should be mapped
to a single individual::CategoricalVariable. For example, a
model which classifies individuals as in an SIR state and in a
high or low risk groups could be represented with two categorical
variable objects
There can be an unlimited number of categorical variable objects, but
every individual can only take on a single value for each of them at any
time. The allowable (categorical) state space for each individual is a
contingency table where the margins are given by the values for each
individual::CategoricalVariable.
individual::CategoricalVariable objects internally store
state as a hash table of category values (with string keys), each of
which contains a individual::Bitset recording the
individuals in that state, making operations on
individual::CategoricalVariable objects, or chains of
operations extremely fast, and therefore should be the preferred
variable type for discrete variables. However, cases in which discrete
variables have a large number of values or when many values have few or
no individuals in that category should use the
individual::IntegerVariable instead.
Integer Variable
An individual::IntegerVariable should be used for
discrete variables that may either be technically unbounded, or whose
set of possible values is so large that a
individual::CategoricalVariable is impractical.
individual::IntegerVariable objects internally stores state
as a vector of integers of length equal to the number of individuals in
the simulation, to be contrasted with the hash table and bitsets used in
individual::CategoricalVariable. Examples of this variable
type might include household for a model that simulated transmission of
disease between a community of households, or age group for an
age-structured model.
Double Variable
A individual::DoubleVariable can be used for continuous
variables that are represented as double precision floating-point
numbers (hence, “double”). Such variables are internally stored as a
vector of doubles of length equal to the number of individuals in the
simulation. Examples of this variable type might include levels of
immune response or concentration of some environmental pollutant in the
blood.
Processes
In “individual”, processes are functions which are called on each
time step with a single argument, t, the current time step
of the simulation. Processes are how most of the dynamics of a model are
specified, and are implemented as closures,
which are functions that are able to access data not included in the
list of function arguments.
An illustrative example can be found in the provided
individual::bernoulli_process, which moves individuals from
one value (source) of a individual::CategoricalVariable to
another (destination) at a constant probability (such that the dwell
time in the from state follows a geometric
distribution).
bernoulli_process <- function(variable, from, to, rate) {
function(t) {
variable$queue_update(
to,
variable$get_index_of(from)$sample(rate)
)
}
}We can test empirically that dwell times do indeed follow a geometric
distribution (and demonstrate uses of
individual::IntegerVariable objects) by modifying slightly
the function to take int_variable, an object of class
individual::IntegerVariable. Each time the returned
function closure is called it randomly samples a subset of individuals
to move to the destination state, and records the time step at which
those individuals move in the individual::IntegerVariable.
We run the simple simulation loop until there are no more individuals
left in the source state, and compare the dwell times in the source
state to the results of stats::rgeom, and see that they are
from the same distribution, using stats::chisq.test for
comparison of discrete distributions.
bernoulli_process_time <- function(variable, int_variable, from, to, rate) {
function(t) {
to_move <- variable$get_index_of(from)$sample(rate)
int_variable$queue_update(values = t, index = to_move)
variable$queue_update(to, to_move)
}
}
n <- 5e4
state <- CategoricalVariable$new(c('S', 'I'), rep('S', n))
time <- IntegerVariable$new(initial_values = rep(0, n))
proc <- bernoulli_process_time(variable = state,int_variable = time,from = 'S',to = 'I',rate = 0.1)
t <- 0
while (state$get_size_of('S') > 0) {
proc(t = t)
state$.update()
time$.update()
t <- t + 1
}
times <- time$get_values()
chisq.test(times, rgeom(n,prob = 0.1),simulate.p.value = T)
#>
#> Pearson's Chi-squared test with simulated p-value (based on 2000
#> replicates)
#>
#> data: times and rgeom(n, prob = 0.1)
#> X-squared = 7243.4, df = NA, p-value = 0.3503By using individual::IntegerVariable and
individual::DoubleVariable objects to modify the
probability with which individuals move between states, as well as
counters for recording previous state transitions and times, complex
dynamics can be specified from simple building blocks. Several “prefab”
functions are provided to make function closures corresponding to state
transition processes common in epidemiological models.
Render
During each time step we often want to record (render) certain output from the simulation. Because rendering output occurs each time step, they are called as other processes during the simulation loop (this is another way we could have implemented the time counters in the previous example).
In “individual”, rendering output is handled by
individual::Render class objects, which build up the
recorded data and can return a base::data.frame object for
plotting and analysis. A individual::Render object is then
stored in a function closure that is called each time step like other
processes, and into which data may be recorded. Please note that the
function closure must also store any variable objects whose values
should be tracked, such as the prefab rendering process
individual::categorical_count_renderer_process, taking a
individual::Render object as the first argument and a
individual::CategoricalVariable object as the second.
categorical_count_renderer_process <- function(renderer, variable, categories) {
function(t) {
for (c in categories) {
renderer$render(paste0(c, '_count'), variable$get_size_of(c), t)
}
}
}Events
While technically nothing more than variables and processes is
required to simulate complex models in “individual”, keeping track of
auxiliary state, such as counters which track the amount of time until
something happens (e.g., a non-geometric waiting time distribution) can
quickly add unnecessary frustration and complexity. The
individual::Event class is a way to model events that don’t
occur every time step, but are instead scheduled to fire at some future
time after a delay. The scheduled events can be canceled, if something
occurs first that should preempt them.
Events can be scheduled by processes; when an event fires, the event
listener is called. The event listener is an arbitrary function that is
called with the current time step t as its sole argument.
Listeners can execute any state changes, and can themselves schedule
future events (not necessarily of the same type). Listeners are
implemented as function closures, just like processes. Events can have any number of listeners
attached to them which are called when the event fires.
Targeted Events
Unlike individual::Event objects, whose listeners are
called with a single argument t, when the listeners of
individual::TargetedEvent objects are called, two arguments
are passed. The first is the current time step t, and the
second is a individual::Bitset object giving the indies of
individuals which will be affected by the event. Because it stores these
objects internally to schedule future updates, initialization of
individual::TargetedEvent objects requires the total
population size as an argument. When scheduling a
individual::TargetedEvent, a user provides a vector or
individual::Bitset of indicies for those individuals which
will have the event scheduled. In addition, a user provides a delay
which the event will occur after. The delay can either be a single
value, so all individuals will experience the event after that delay, or
a vector of values equal to the number of individuals who were
scheduled. This allows, for example individual heterogeneity in
distribution of waiting times for some event.
Much like regular Events, previously scheduled
targeted events can be canceled, requiring either a
individual::Bitset or vector of indices for individuals
whose events should be canceled.
The code to define events is typically quite brief, although
typically another process must be defined that queues them. A recovery
event in an SIR model for example, might look like this, where
state is a
individual::CategoricalVariable:
recovery_event <- TargetedEvent$new(population_size = 100)
recovery_event$add_listener(function(timestep, target) {
state$queue_update('R', target)
})Simulate
Using a combination of variables, processes, and events, highly
complex individual based models can be specified, and simulated with the
function individual::simulation_loop, which defines the
order in which state updates are preformed.
It is possible to create conflicts between different processes, for example, if a single individual is subject to a death process and infection process, then both death and infection state changes could be scheduled for that individual during a time step. When processes or events create conflicting state updates, the last update will take precedence. Another way to make sure conflicts do not exist is to ensure only a single process is called for each state which samples each individual’s update among all possible updates which could happen for those individuals, so that conflicting updates cannot be scheduled for individuals in a state.
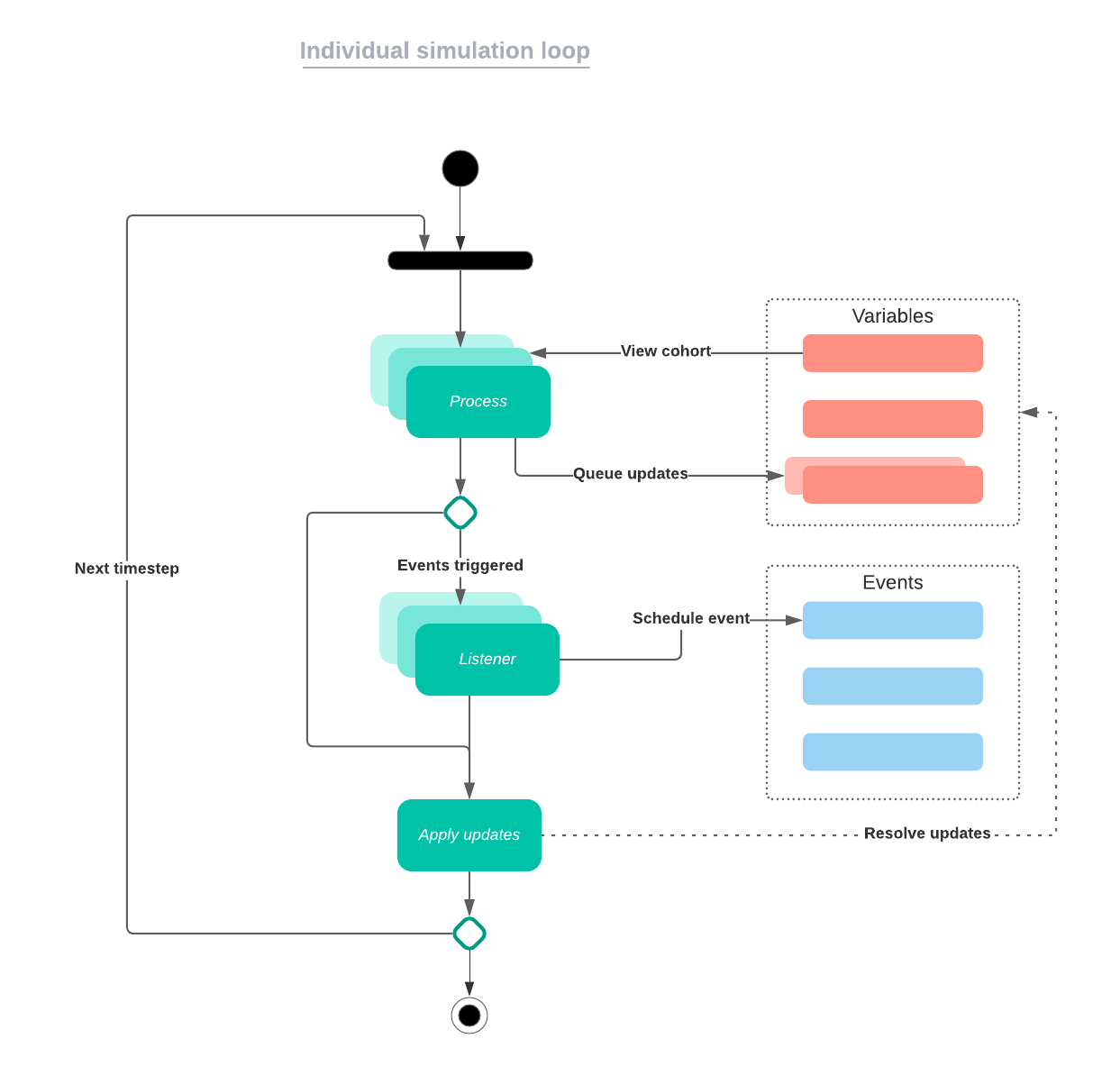
Simulation loop flowchart
The equivalent code used in the simulation loop is below.
simulation_loop <- function(
variables = list(),
events = list(),
processes = list(),
timesteps
) {
if (timesteps <= 0) {
stop('End timestep must be > 0')
}
for (t in seq_len(timesteps)) {
for (process in processes) {
execute_any_process(process, t)
}
for (event in events) {
event$.process()
}
for (variable in variables) {
variable$.update()
}
for (event in events) {
event$.tick()
}
}
}