More Flexible Model Run
H. Juliette T. Unwin & Joel Hellewell
2025-02-05
run_model.RmdIntroduction
If you are able to the run the run_model_example
function (see this vignette),
then you can move on to running the base malaria model more flexibly or
run alternative models with different intervention methods.
Default use of function
The run_model function has the following default
arguments:
- model - The name of the malaria model to run (default = base model)
- The number of heterogeneity brackets -
het_brackets = 5 - The vector of age compartments -
age = c(0,0.25,0.5,0.75,1,1.25,1.5,1.75,2,3.5,5,7.5,10,15,20,30,40,50,60,70,80) - The initial EIR -
init_EIR = 10 - The percentage of the population that gets treated -
init_ft = 0.4 - The country -
country = NULL - The admin 2 unit -
admin2 = NULL
By default the run_model function runs the model for 100
days in a country with no seasonality (achieved by setting
admin2 and country equal to NULL)
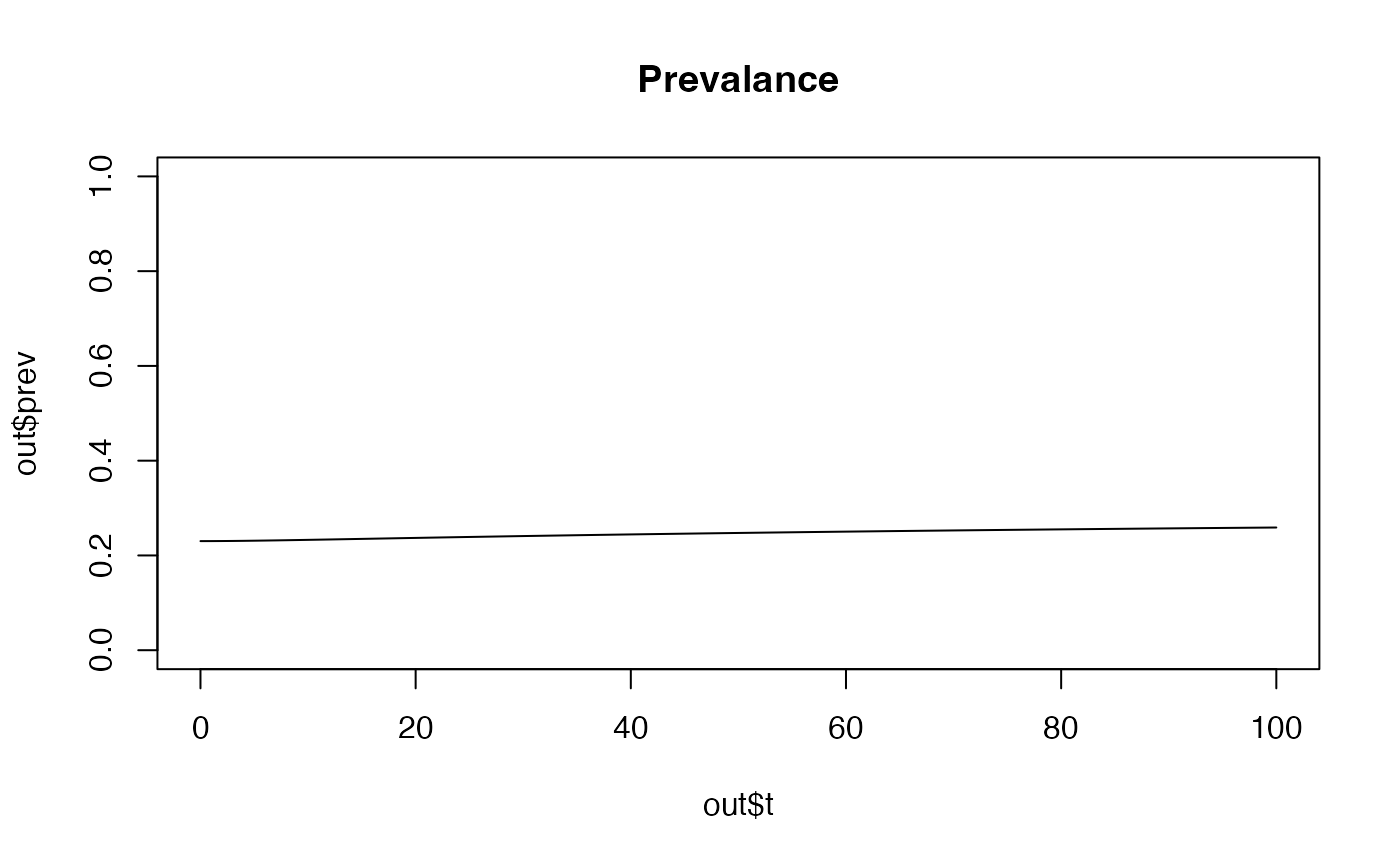
and no interventions. We can run the model and plot the prevalence as
follows:
library(ICDMM)
out <- run_model()
plot(out$t,out$prev, main= "Prevalance", ylim = c(0, 1), type='l')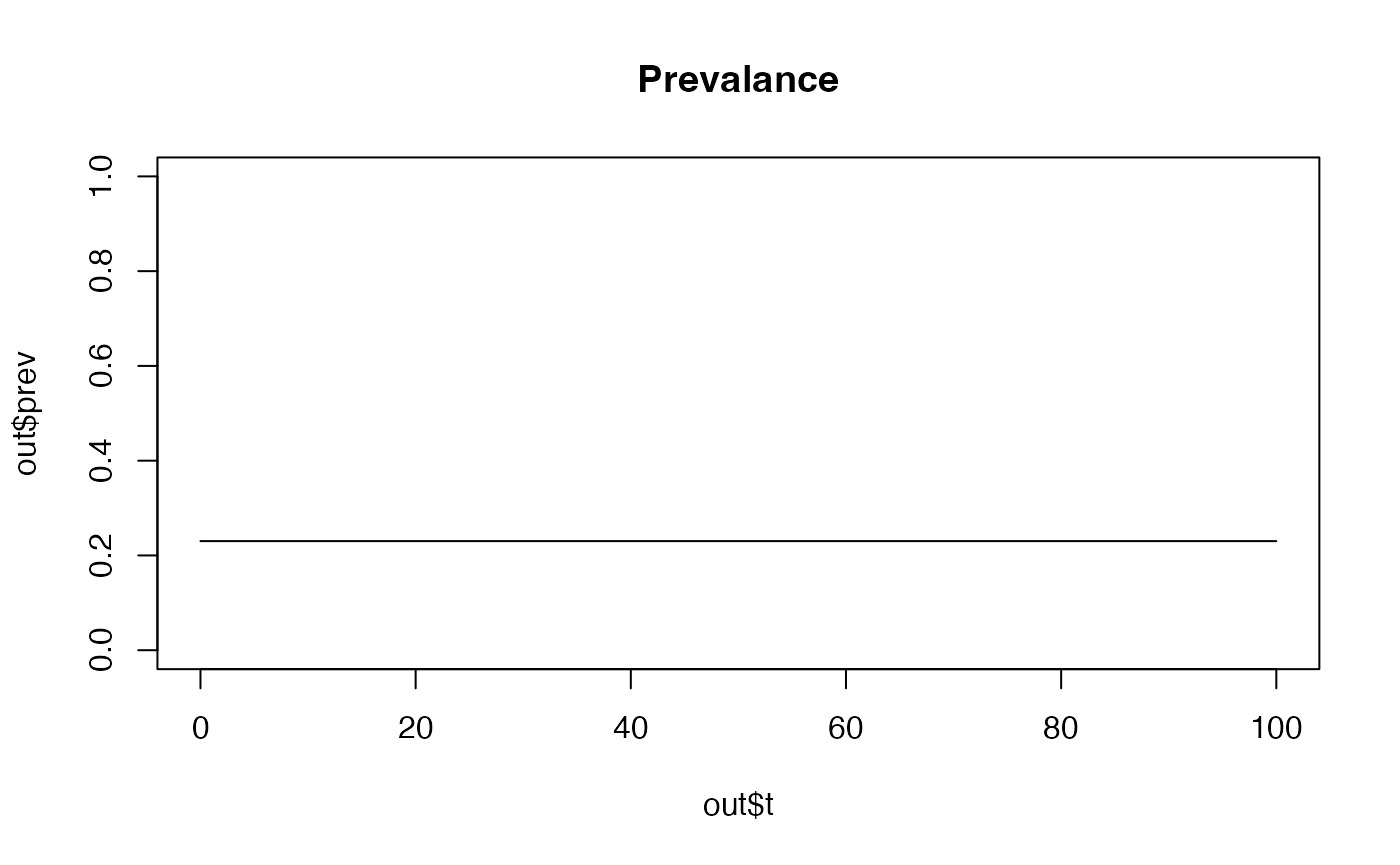
Providing alternative arguments
You can provide new parameter values for parameters in the model
(model parameters are listed here) by including them as arguments in the
run_model function:
It is possible to change the default arguments for the
create_r_model function as shown below by setting the
initial EIR (the average number of infectious bites per person each
year) to 50 instead of the default value of 10:
We can also change the length of time that the model is run for by
setting time = 1000 (days) and introduce an intervention by
giving 75% of the population an insecticidal bed net
(itn_cov = 0.75) after 365 days
(ITN_IRS_on = 365). We also need to specify the number of
intervention categories (num_int), if we introduce nets we
need 2 categories for people with and without nets. If we introduce ITNs
and IRS, we need 4 categories for no intervention, nets only, IRS only,
and both nets and IRS. Here we compare introducing 75% ITN coverage
after 365 days to 75% ITN coverage + 40% IRS coverage:
# ITN only
out <- run_model(init_EIR = 50,
time = 1000,
ITN_IRS_on = 365,
itn_cov = 0.75,
num_int = 2)
# ITN + IRS
out2 <- run_model(init_EIR = 50,
time = 1000,
ITN_IRS_on = 365,
itn_cov = 0.75,
irs_cov = 0.4,
num_int = 4)
plot(out$t,out$prev, main= "Prevalance", type='l', ylim = c(0, 1))
lines(out2$t, out2$prev, col = "blue")
abline(v = 365, lty = 2)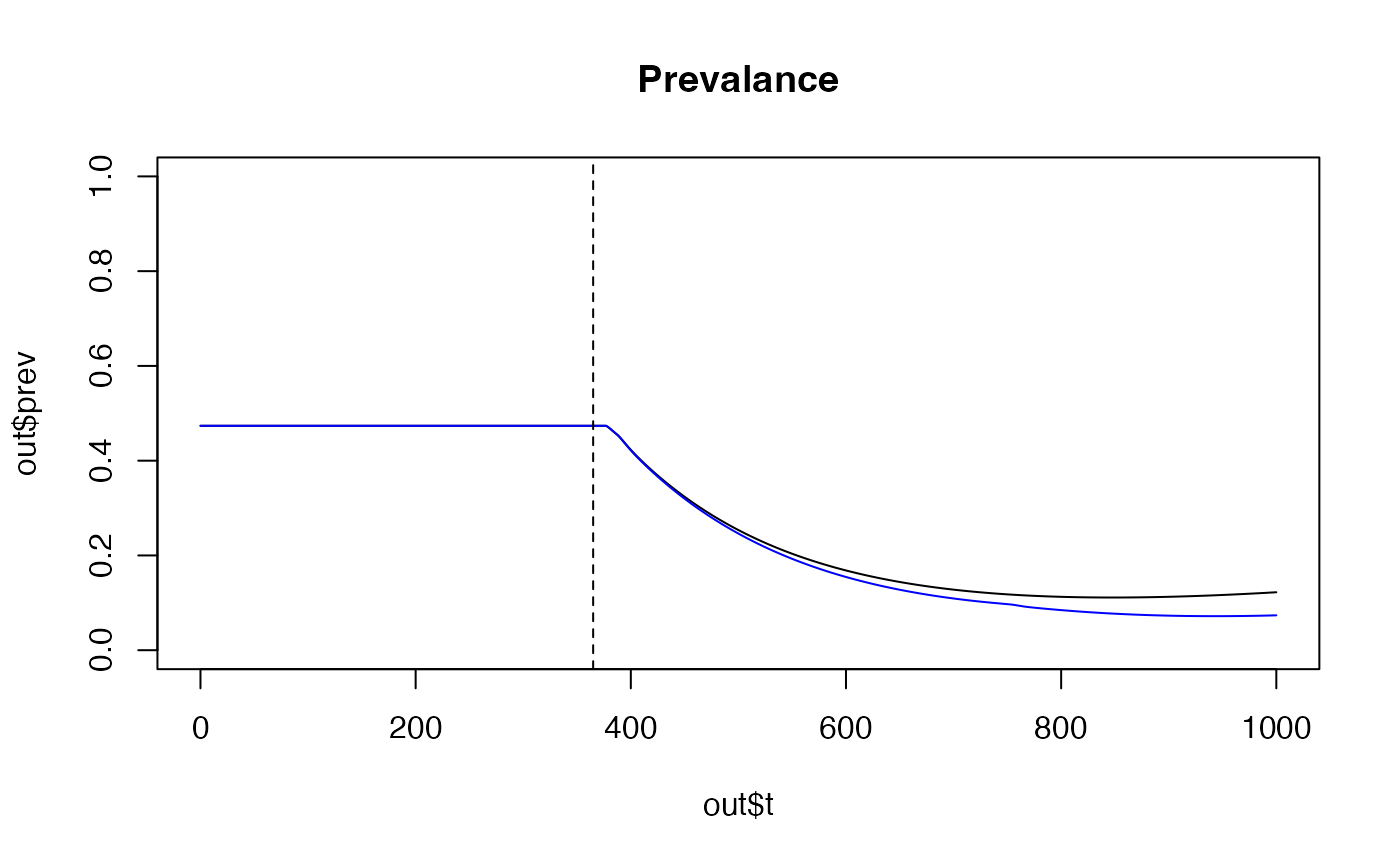
Changing seasonality parameters
It is possible to change the seasonality parameters by specifying a
named admin unit or both an admin unit and country to the
run_model function. The latter is better when the admin
name is common. Fuzzy matching is used to match the provided arguments
with the data set provided in the package. The data set can be loaded
into your workspace as follows:
seasonality_data <- ICDMM::load_file("admin_units_seasonal.rds")You can then specify the location that you would like to load the
seasonality profile for by specifying country and
admin2 using character strings when calling the
run_model function (An example is shown in the previous vignette.
Using alternative models
It is also possible to change the odin model by editing the
model argument. By default the run_model
function uses the basic malaria model (called odin_model).
You can see the four different models currently included in the package
in the inst/odin directory. To run a different model just
specify its name, however you may encounter errors unless you specify
parameter values for extra parameters in the alternative models that may
not have a default value.
## Running this will give an error because we have not specified the value of the parameter hrp2_prop
#run_model(model = "odin_model_hrp2")
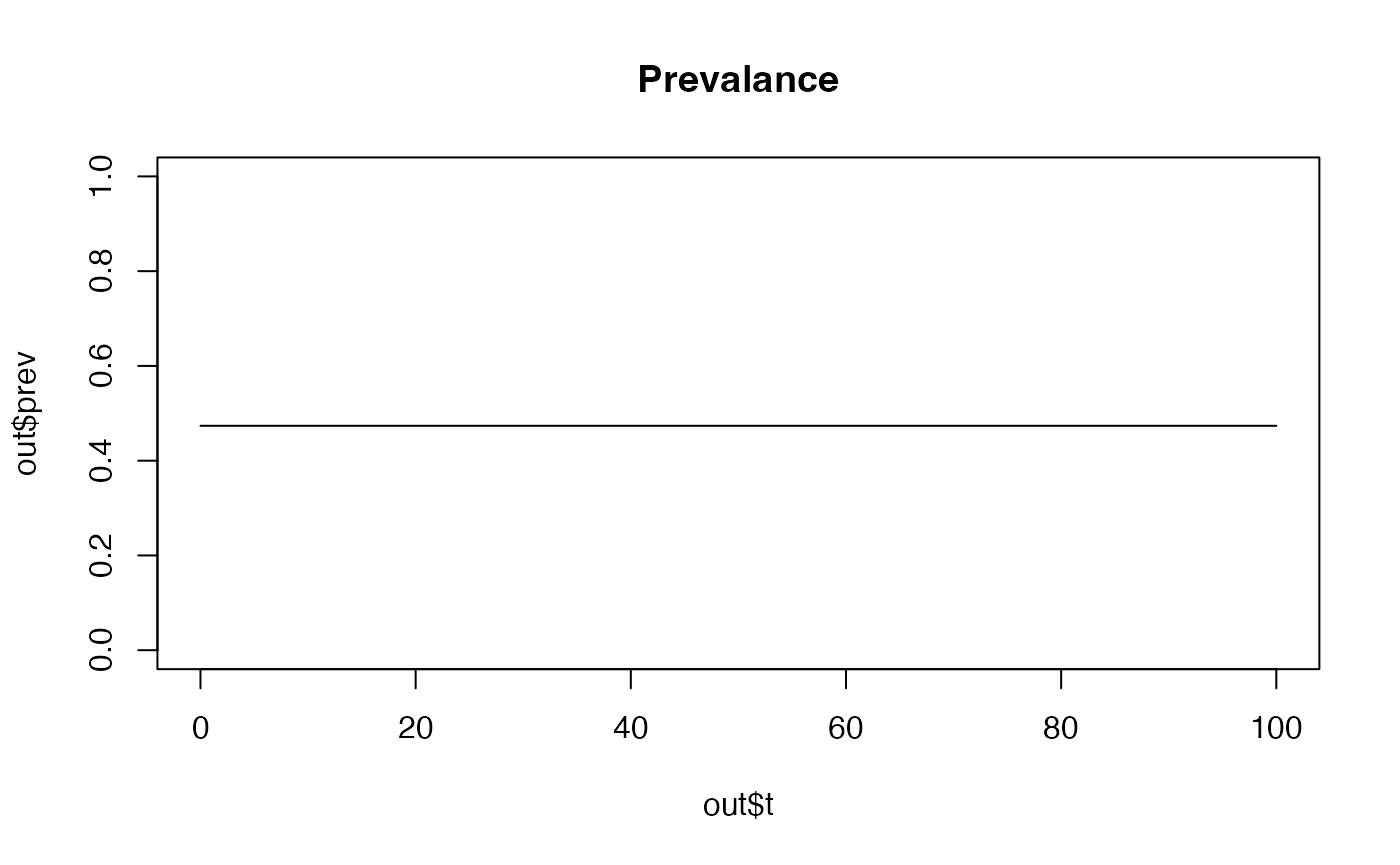
## We can specify the new parameter value in the function call
out <- run_model(model = "odin_model_hrp2", hrp2_prop = 0.5)
plot(out$t,out$prev, main= "Prevalance", type='l', ylim = c(0, 1))