Overall Infectivity Due To Previously Infected Individuals
Source:R/overall_infectivity.R
overall_infectivity.Rdoverall_infectivity computes the overall infectivity due to previously
infected individuals.
overall_infectivity(incid, si_distr)Arguments
- incid
One of the following
A vector (or a dataframe with a single column) of non-negative integers containing an incidence time series
A dataframe of non-negative integers with two columns, so that
incid$localcontains the incidence of cases due to local transmission andincid$importedcontains the incidence of imported cases (withincid$local + incid$importedthe total incidence).
Note that the cases from the first time step are always all assumed to be imported cases.
- si_distr
Vector of probabilities giving the discrete distribution of the serial interval.
Value
A vector which contains the overall infectivity \(\lambda_t\) at each time step
Details
The overall infectivity \(\lambda_t\) at time step \(t\) is
equal to the sum of the previously infected individuals (given by the
incidence vector \(I\), with I = incid$local + incid$imported if
\(I\) is a matrix), weigthed by their infectivity at time \(t\) (given by
the discrete serial interval distribution \(w_k\)). In mathematical terms:
\(\lambda_t = \sum_{k=1}^{t-1}I_{t-k}w_k\)
References
Cori, A. et al. A new framework and software to estimate time-varying reproduction numbers during epidemics (AJE 2013).
See also
Examples
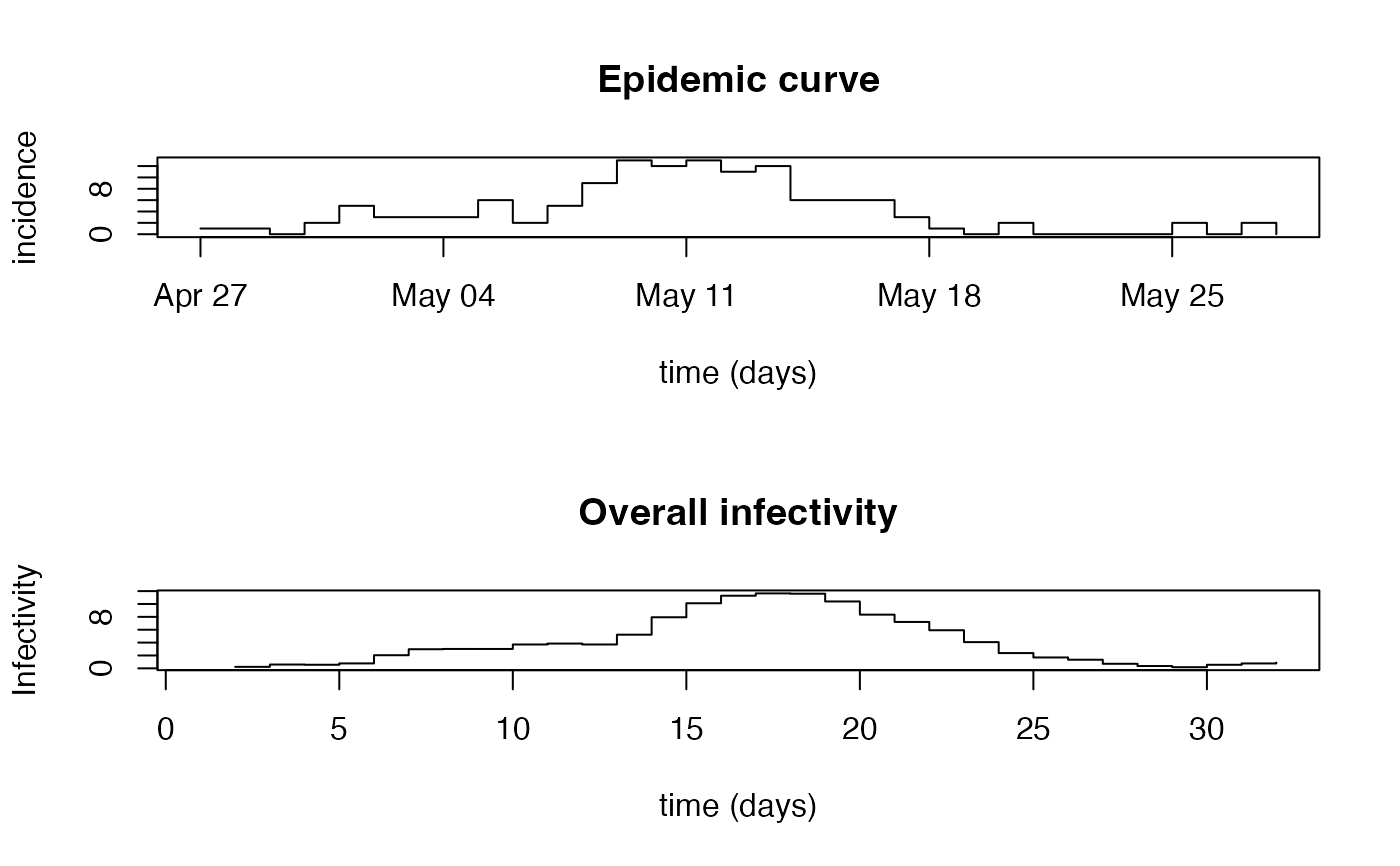
## load data on pandemic flu in a school in 2009
data("Flu2009")
## compute overall infectivity
lambda <- overall_infectivity(Flu2009$incidence, Flu2009$si_distr)
par(mfrow=c(2,1))
plot(Flu2009$incidence, type = "s", xlab = "time (days)", ylab = "incidence")
title(main = "Epidemic curve")
plot(lambda, type = "s", xlab = "time (days)", ylab = "Infectivity")
title(main = "Overall infectivity")