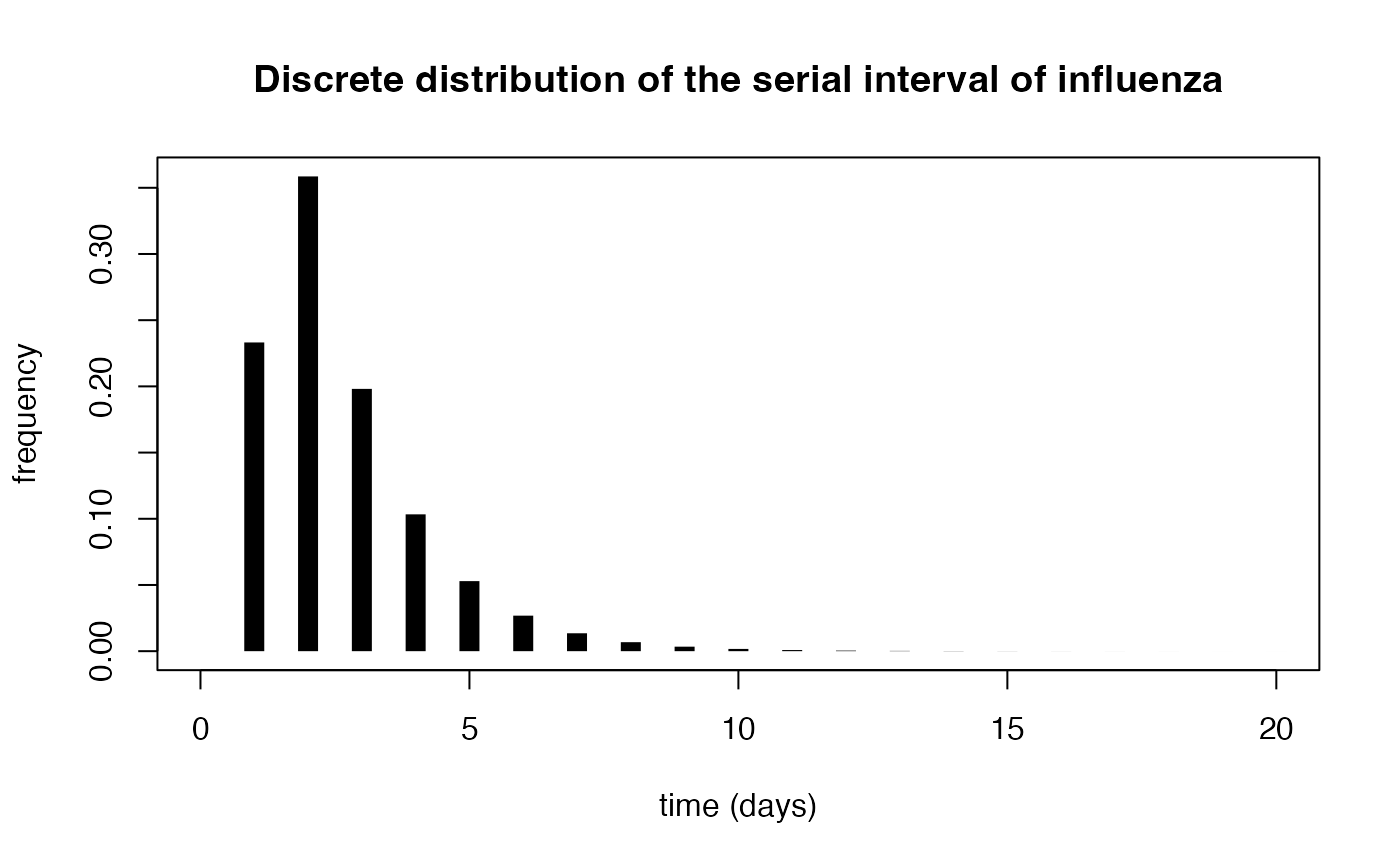
Discretized Generation Time Distribution Assuming A Shifted Gamma Distribution
Source:R/discr_si.R
discr_si.Rddiscr_si computes the discrete distribution of the serial interval,
assuming that the serial interval is shifted Gamma distributed, with shift 1.
discr_si(k, mu, sigma)Arguments
Value
Gives the discrete probability \(w_k\) that the serial interval is equal to \(k\).
Details
Assuming that the serial interval is shifted Gamma distributed with mean \(\mu\), standard deviation \(\sigma\) and shift \(1\), the discrete probability \(w_k\) that the serial interval is equal to \(k\) is:$$w_k = kF_{\{\mu-1,\sigma\}}(k)+(k-2)F_{\{\mu-1,\sigma\}} (k-2)-2(k-1)F_{\{\mu-1,\sigma\}}(k-1)\\ +(\mu-1)(2F_{\{\mu-1+\frac{\sigma^2}{\mu-1}, \sigma\sqrt{1+\frac{\sigma^2}{\mu-1}}\}}(k-1)- F_{\{\mu-1+\frac{\sigma^2}{\mu-1}, \sigma\sqrt{1+\frac{\sigma^2}{\mu-1}}\}}(k-2)- F_{\{\mu-1+\frac{\sigma^2}{\mu-1}, \sigma\sqrt{1+\frac{\sigma^2}{\mu-1}}\}}(k))$$where \(F_{\{\mu,\sigma\}}\) is the cumulative density function of a Gamma distribution with mean \(\mu\) and standard deviation \(\sigma\).
References
Cori, A. et al. A new framework and software to estimate time-varying reproduction numbers during epidemics (AJE 2013).
See also
Examples
## Computing the discrete serial interval of influenza
mean_flu_si <- 2.6
sd_flu_si <- 1.5
dicrete_si_distr <- discr_si(seq(0, 20), mean_flu_si, sd_flu_si)
plot(seq(0, 20), dicrete_si_distr, type = "h",
lwd = 10, lend = 1, xlab = "time (days)", ylab = "frequency")
title(main = "Discrete distribution of the serial interval of influenza")