This data set gives:
the UK daily incidence of deaths within 28 days of a positive COVID test (see source and references),
the discrete daily distribution of the serial interval for SARS-CoV-2, assuming a shifted Gamma distribution with mean 6.1 days, standard deviation 4.2 days and shift 1 day (see references).
Format
A list of two elements:
incidence: a data.frame containing 672 days of observation,
si_distr: a vector containing a set of 30 probabilities.
Source
Bi Q, Wu Y, Mei S, Ye C, Zou X, Zhang Z, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. The Lancet Infectious Diseases. 2020 Aug 1;20(8):911–9.
https://coronavirus.data.gov.uk/details/cases
References
Nash RK, Cori A, Nouvellet P. Estimating the epidemic reproduction number from temporally aggregated incidence data: a statistical modelling approach and software tool. (medRxiv pre-print)
Bi Q, Wu Y, Mei S, Ye C, Zou X, Zhang Z, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. The Lancet Infectious Diseases. 2020 Aug 1;20(8):911–9.
Examples
data("covid_deaths_2020_uk")
## estimate the reproduction number (method "non_parametric_si")
res <- estimate_R(covid_deaths_2020_uk$incidence$Incidence,
method="non_parametric_si",
config = make_config(list(si_distr = covid_deaths_2020_uk$si_distr)))
#> Default config will estimate R on weekly sliding windows.
#> To change this change the t_start and t_end arguments.
plot(res)
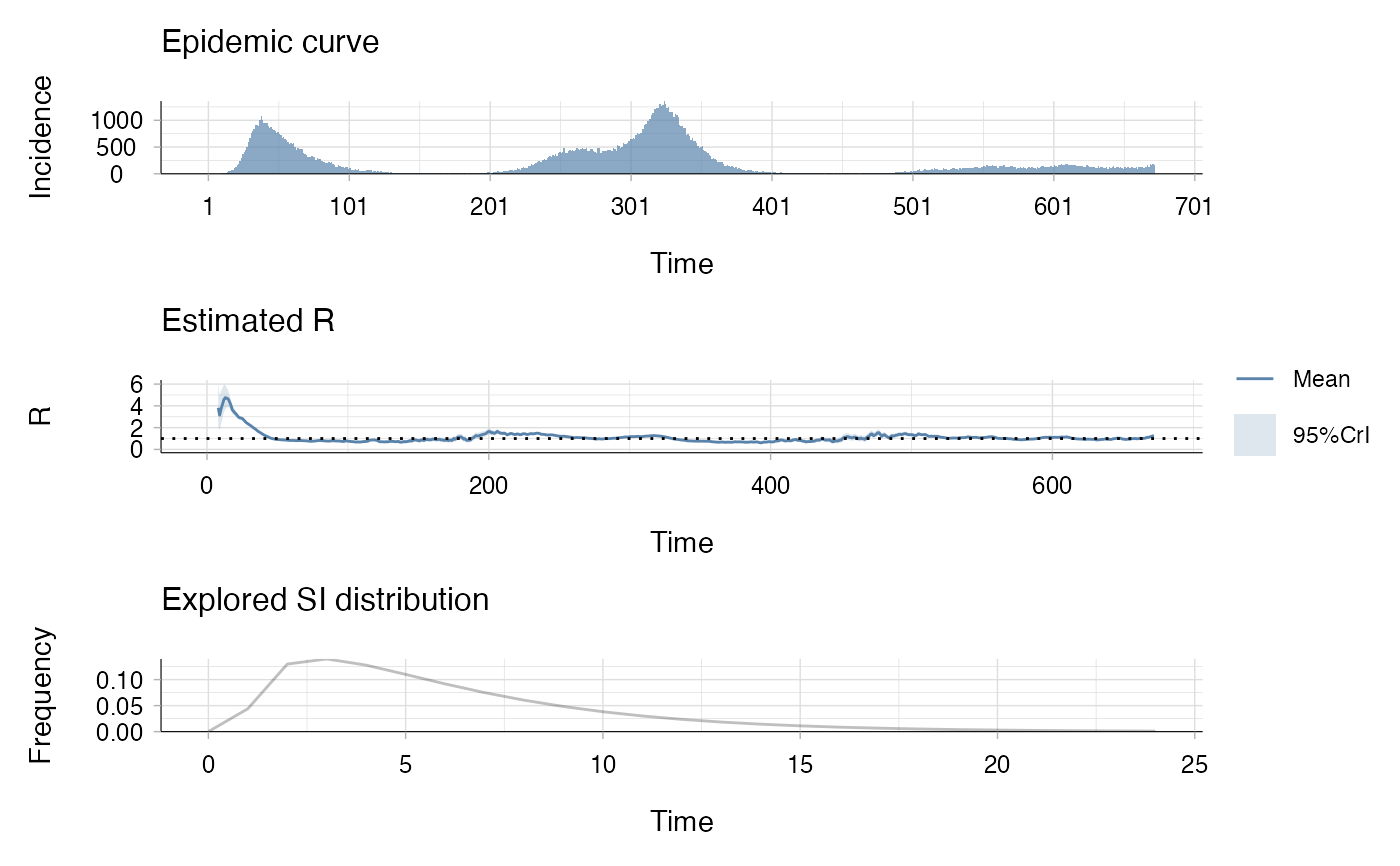 ## the second plot produced shows, at each each day,
## the estimate of the reproduction number
## over the 7-day window finishing on that day.
## the second plot produced shows, at each each day,
## the estimate of the reproduction number
## over the 7-day window finishing on that day.