This data set gives:
the daily incidence of onset of disease in Baltimore during the 1918 H1N1 influenza pandemic (see source and references),
the discrete daily distribution of the serial interval for influenza, assuming a shifted Gamma distribution with mean 2.6 days, standard deviation 1.5 days and shift 1 day (see references).
Format
A list of two elements:
incidence: a vector containing 92 days of observation,
si_distr: a vector containing a set of 12 probabilities.
Source
Frost W. and E. Sydenstricker (1919) Influenza in Maryland: preliminary statistics of certain localities. Public Health Rep.(34): 491-504.
References
Cauchemez S. et al. (2011) Role of social networks in shaping disease transmission during a community outbreak of 2009 H1N1 pandemic influenza. Proc Natl Acad Sci U S A 108(7), 2825-2830.
Ferguson N.M. et al. (2005) Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 437(7056), 209-214.
Fraser C. et al. (2011) Influenza Transmission in Households During the 1918 Pandemic. Am J Epidemiol 174(5): 505-514.
Frost W. and E. Sydenstricker (1919) Influenza in Maryland: preliminary statistics of certain localities. Public Health Rep.(34): 491-504.
Vynnycky E. et al. (2007) Estimates of the reproduction numbers of Spanish influenza using morbidity data. Int J Epidemiol 36(4): 881-889.
White L.F. and M. Pagano (2008) Transmissibility of the influenza virus in the 1918 pandemic. PLoS One 3(1): e1498.
Examples
## load data on pandemic flu in Baltimore in 1918
data("Flu1918")
## estimate the reproduction number (method "non_parametric_si")
res <- estimate_R(Flu1918$incidence,
method = "non_parametric_si",
config = make_config(list(si_distr = Flu1918$si_distr)))
#> Default config will estimate R on weekly sliding windows.
#> To change this change the t_start and t_end arguments.
plot(res)
#> Warning: The `guide` argument in `scale_*()` cannot be `FALSE`. This was deprecated in
#> ggplot2 3.3.4.
#> ℹ Please use "none" instead.
#> ℹ The deprecated feature was likely used in the incidence package.
#> Please report the issue at <https://github.com/reconhub/incidence/issues>.
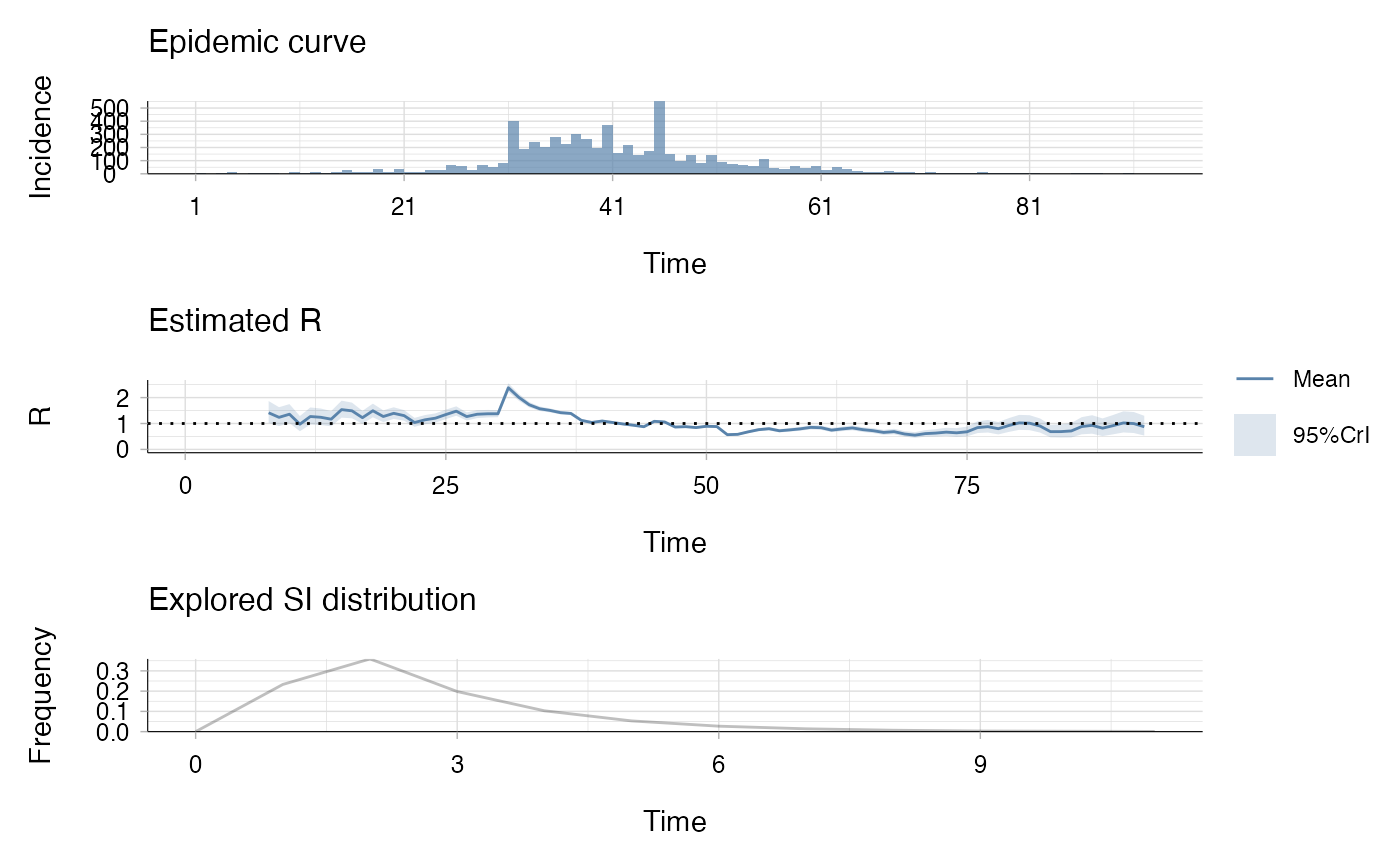 ## the second plot produced shows, at each each day,
## the estimate of the reproduction number
## over the 7-day window finishing on that day.
## the second plot produced shows, at each each day,
## the estimate of the reproduction number
## over the 7-day window finishing on that day.