library(orderly)
orderly_init("workdir/part2")
## ✔ Created orderly root at '/home/runner/work/orderly-tutorial/orderly-tutorial/workdir/part2'
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`Orderly Code
What can I do?
- orderly code is any code you can use from R
- Use (almost) any package, any sort of file
But…
- The directory above your file does not exist
- Don’t use absolute paths or
../path fragments - You can add metadata to help future you/others
A clean beginning
Suppose we’re working on a data analysis pipeline, starting with “incoming data”:
orderly_new("incoming")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ✔ Created 'src/incoming/incoming.R'Our setup:
Incoming data
I’ve copied some data in as data.xlsx into src/incoming.
- Modify
incoming.Rto tidy that up for consumption using your favourite packages. - Set your working directory to
src/incomingand just edit things as usual - Which sheet contains the data?
- Where is the data in that sheet?
- Do you like those column names?
- How about that date format?
Incoming data, cleaned
My attempt at cleaning:
- Read in the data
- Clean up the names (could have used
janitor) - Convert date format
- Write as
csv
Incoming data, running things
id <- orderly_run("incoming")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'incoming' `20251007-071623-1226e67d` at 2025-10-07 07:16:23.079066
## > d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
## > names(d) <- gsub(" ", "_", tolower(names(d)))
## > d$date <- as.Date(d$date)
## > write.csv(d, "data.csv", row.names = FALSE)
## ✔ Finished running 'incoming.R'
## ℹ Finished 20251007-071623-1226e67d at 2025-10-07 07:16:23.149212 (0.07014561 secs)Our generated metadata (this output box scrolls)
orderly_metadata(id)
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## $schema_version
## [1] "0.1.1"
##
## $name
## [1] "incoming"
##
## $id
## [1] "20251007-071623-1226e67d"
##
## $time
## $time$start
## [1] "2025-10-07 07:16:23 UTC"
##
## $time$end
## [1] "2025-10-07 07:16:23 UTC"
##
##
## $parameters
## NULL
##
## $files
## path size
## 1 data.csv 466
## 2 data.xlsx 10851
## 3 incoming.R 174
## hash
## 1 sha256:9117700f079b786812cc20904f4c34f5659f2986a923d2771216551cf378e86f
## 2 sha256:149445ecc545eb987ecc0d5255a48f21165ee1c9b6b54c84b882ce1fbda066b7
## 3 sha256:8ce9dc59614d62b39cd4ff8cfba08e48f993a9be6d3c29357118df386bf7688f
##
## $depends
## [1] packet query files
## <0 rows> (or 0-length row.names)
##
## $git
## $git$sha
## [1] "6ddafeaae72937fa2cceb1817b8d133b64d5293c"
##
## $git$branch
## [1] "main"
##
## $git$url
## [1] "https://github.com/mrc-ide/orderly-tutorial"
##
##
## $custom
## $custom$orderly
## $custom$orderly$artefacts
## [1] description paths
## <0 rows> (or 0-length row.names)
##
## $custom$orderly$role
## path role
## 1 incoming.R orderly
##
## $custom$orderly$description
## $custom$orderly$description$display
## NULL
##
## $custom$orderly$description$long
## NULL
##
## $custom$orderly$description$custom
## NULL
##
##
## $custom$orderly$shared
## [1] here there
## <0 rows> (or 0-length row.names)
##
## $custom$orderly$session
## $custom$orderly$session$platform
## $custom$orderly$session$platform$version
## [1] "R version 4.5.1 (2025-06-13)"
##
## $custom$orderly$session$platform$os
## [1] "Ubuntu 24.04.3 LTS"
##
## $custom$orderly$session$platform$system
## [1] "x86_64, linux-gnu"
##
##
## $custom$orderly$session$packages
## package version attached
## 1 orderly 1.99.92 TRUE
## 2 crayon 1.5.3 FALSE
## 3 vctrs 0.6.5 FALSE
## 4 cli 3.6.5 FALSE
## 5 knitr 1.50 FALSE
## 6 rlang 1.1.6 FALSE
## 7 xfun 0.53 FALSE
## 8 jsonlite 2.0.0 FALSE
## 9 glue 1.8.0 FALSE
## 10 openssl 2.3.4 FALSE
## 11 askpass 1.2.1 FALSE
## 12 htmltools 0.5.8.1 FALSE
## 13 sys 3.4.3 FALSE
## 14 readxl 1.4.5 FALSE
## 15 rmarkdown 2.30 FALSE
## 16 cellranger 1.1.0 FALSE
## 17 evaluate 1.0.5 FALSE
## 18 tibble 3.3.0 FALSE
## 19 fastmap 1.2.0 FALSE
## 20 yaml 2.3.10 FALSE
## 21 lifecycle 1.0.4 FALSE
## 22 compiler 4.5.1 FALSE
## 23 fs 1.6.6 FALSE
## 24 pkgconfig 2.0.3 FALSE
## 25 digest 0.6.37 FALSE
## 26 gert 2.1.5 FALSE
## 27 R6 2.6.1 FALSE
## 28 pillar 1.11.1 FALSE
## 29 credentials 2.0.3 FALSE
## 30 magrittr 2.0.4 FALSE
## 31 withr 3.0.2 FALSE
## 32 tools 4.5.1 FALSE“Resources”
- Any file that is an input
- For example:
- Scripts that you
source() - R Markdown files for
knitrorrmarkdown - Data files (
.csv,.xlsx, etc) - Plain text files (
README.md, licence info, etc)
- Scripts that you
- Here,
data.xlsxis an input
Telling orderly about resources
- Tells orderly
data.xlsxis a resource - Fail early if resource not found
- Error if resource is modified
- Extra metadata, advertising what files were used
id <- orderly_run("incoming")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'incoming' `20251007-071623-3df771be` at 2025-10-07 07:16:23.246878
## > orderly_resource("data.xlsx")
## > d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
## > names(d) <- gsub(" ", "_", tolower(names(d)))
## > d$date <- as.Date(d$date)
## > write.csv(d, "data.csv", row.names = FALSE)
## ✔ Finished running 'incoming.R'
## ℹ Finished 20251007-071623-3df771be at 2025-10-07 07:16:23.283559 (0.03668118 secs)
orderly_metadata(id)$custom$orderly$role
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## path role
## 1 incoming.R orderly
## 2 data.xlsx resource“Artefacts”
- Any file that is an output
- For example:
- Datasets you generate
- html or pdf output from
knitrorrmarkdown - Plain text files
- Inputs themselves, sometimes
- Here,
data.csvis an artefact
Telling orderly about artefacts
- Tells orderly
csv.xlsxis an artefact - Fail if artefact not produced
- Extra metadata, advertising what files were produced
id <- orderly_run("incoming")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'incoming' `20251007-071623-5dae8f81` at 2025-10-07 07:16:23.370757
## > orderly_resource("data.xlsx")
## > orderly_artefact(files = "data.csv", description = "Cleaned data")
## > d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
## > names(d) <- gsub(" ", "_", tolower(names(d)))
## > d$date <- as.Date(d$date)
## > write.csv(d, "data.csv", row.names = FALSE)
## ✔ Finished running 'incoming.R'
## ℹ Finished 20251007-071623-5dae8f81 at 2025-10-07 07:16:23.399106 (0.02834916 secs)
orderly_metadata(id)$custom$orderly$artefacts
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## description paths
## 1 Cleaned data data.csvMore metadata
orderly_description(
display = "Incoming data from Otherlandia",
long = "Data as given to us from the MoH in Otherlandia.",
custom = list(received = "2024-10-22"))
orderly_resource("data.xlsx")
orderly_artefact(files = "data.csv", description = "Cleaned data")
d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
names(d) <- gsub(" ", "_", tolower(names(d)))
d$date <- as.Date(d$date)
write.csv(d, "data.csv", row.names = FALSE)Running this:
id <- orderly_run("incoming", echo = FALSE)
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'incoming' `20251007-071623-7af00ffb` at 2025-10-07 07:16:23.487372
## ✔ Finished running 'incoming.R'
## ℹ Finished 20251007-071623-7af00ffb at 2025-10-07 07:16:23.515404 (0.02803135 secs)
orderly_metadata(id)$custom$orderly$description
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## $display
## [1] "Incoming data from Otherlandia"
##
## $long
## [1] "Data as given to us from the MoH in Otherlandia."
##
## $custom
## $custom$received
## [1] "2024-10-22"Dependencies
- This is really the point of orderly
- You can pull in any file from any previously run packet
- You can use queries to select packets to depend on
Our aim: We want to use data.csv in some analysis
orderly_new("analysis")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ✔ Created 'src/analysis/analysis.R'Setting up a dependency
orderly_dependency("incoming", "latest", "data.csv")
orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
description = "Plots of coverage")
d <- read.csv("data.csv")
d$date <- as.Date(d$date)
png("coverage-gf.png")
plot(gf_coverage ~ date, d, type = "l")
dev.off()
png("coverage-bf.png")
plot(bf_coverage ~ date, d, type = "l")
dev.off()This is the only file within our analysis directory:
Running the report
id <- orderly_run("analysis")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'analysis' `20251007-071623-a4bac89a` at 2025-10-07 07:16:23.651198
## > orderly_dependency("incoming", "latest", "data.csv")
## ℹ Depending on incoming @ `20251007-071623-7af00ffb` (via latest(name == "incoming"))
## > orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
## + description = "Plots of coverage")
## > d <- read.csv("data.csv")
## > d$date <- as.Date(d$date)
## > png("coverage-gf.png")
## > plot(gf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## > png("coverage-bf.png")
## > plot(bf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## ✔ Finished running 'analysis.R'
## ℹ Finished 20251007-071623-a4bac89a at 2025-10-07 07:16:23.752157 (0.1009593 secs)The aftermath
fs::dir_tree("workdir/part2")
## workdir/part2
## ├── archive
## │ ├── analysis
## │ │ └── 20251007-071623-a4bac89a
## │ │ ├── analysis.R
## │ │ ├── coverage-bf.png
## │ │ ├── coverage-gf.png
## │ │ └── data.csv
## │ └── incoming
## │ ├── 20251007-071623-1226e67d
## │ │ ├── data.csv
## │ │ ├── data.xlsx
## │ │ └── incoming.R
## │ ├── 20251007-071623-3df771be
## │ │ ├── data.csv
## │ │ ├── data.xlsx
## │ │ └── incoming.R
## │ ├── 20251007-071623-5dae8f81
## │ │ ├── data.csv
## │ │ ├── data.xlsx
## │ │ └── incoming.R
## │ └── 20251007-071623-7af00ffb
## │ ├── data.csv
## │ ├── data.xlsx
## │ └── incoming.R
## ├── draft
## │ ├── analysis
## │ └── incoming
## ├── orderly_config.json
## └── src
## ├── analysis
## │ └── analysis.R
## └── incoming
## ├── data.xlsx
## └── incoming.RSome comments on this
- The
data.csvfile has been copied from the final copy ofincomingintoanalysis - The dependency system works interactively too (try it!)
- The logs indicate how dependency resolution occurred
- Metadata about the dependencies is included:
orderly_metadata(id)$depends
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## packet query files
## 1 20251007-071623-7af00ffb latest(name == "incoming") data.csv....Using specific versions
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
orderly_dependency("incoming", "20251007-071623-3df771be", "data.csv")
orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
description = "Plots of coverage")
d <- read.csv("data.csv")
d$date <- as.Date(d$date)
png("coverage-gf.png")
plot(gf_coverage ~ date, d, type = "l")
dev.off()
png("coverage-bf.png")
plot(bf_coverage ~ date, d, type = "l")
dev.off()Running this
id <- orderly_run("analysis")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'analysis' `20251007-071623-e07b6675` at 2025-10-07 07:16:23.881566
## > orderly_dependency("incoming", "20251007-071623-3df771be", "data.csv")
## ℹ Depending on incoming @ `20251007-071623-3df771be` (via single(id == "20251007-071623-3df771be" && name == "incoming"))
## > orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
## + description = "Plots of coverage")
## > d <- read.csv("data.csv")
## > d$date <- as.Date(d$date)
## > png("coverage-gf.png")
## > plot(gf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## > png("coverage-bf.png")
## > plot(bf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## ✔ Finished running 'analysis.R'
## ℹ Finished 20251007-071623-e07b6675 at 2025-10-07 07:16:23.949836 (0.06827044 secs)with metadata
orderly_metadata(id)$depends
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## packet
## 1 20251007-071623-3df771be
## query files
## 1 single(id == "20251007-071623-3df771be" && name == "incoming") data.csv....A more realistic example
orderly_new("wuenic")
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ✔ Created 'src/wuenic/wuenic.R'A real analysis courtesty of Katy Gaythorpe, using wuenic.xlsx
orderly_resource("wuenic.xlsx")
orderly_artefact(
files = "wuenic.rds",
description = "Tidied WUENIC data")
orderly_artefact(
files = "corr_out.rds",
description = "Output correlations between WUENIC and OFFICIAL coverage")
# -------------------------------------------------------------------------
library(dplyr)
library(ggplot2)
library(readxl)
library(janitor)
df <- read_xlsx("wuenic.xlsx")
# Question: is there better agreement in coverage estimates for BCG or YF
# some cleaning
df <- df %>% clean_names()
df <- df %>% mutate(across(starts_with("x"),
.fns = function(inp) as.numeric(gsub("%", "", inp))))
df <- df %>% mutate(all_na_coverage = if_all(starts_with("x"), is.na))
# quick visual for 2023
df %>%
filter(!is.na(antigen)) %>%
filter(country_region %in% c("Nigeria", "Senegal", "Kenya", "Ghana")) %>%
ggplot() +
aes(x = country_region, y = x2023, fill = category) +
geom_col(position = "dodge") +
facet_wrap(antigen ~ ., ncol = 1) +
theme_minimal() +
labs(x = "Country", y = "Coverage in 2023", fill = "Coverage type")
# get correlations per vaccine and country
get_ma_corr <- function(df, country_reg_in = "Afghanistan",
antigen_in = "Yellow fever vaccine") {
df_subset <- df %>% filter(country_region %in% country_reg_in,
antigen %in% antigen_in)
cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", names(df))]),
t(df_subset[df_subset$category %in% "OFFICIAL", grep("^x", names(df))]),
use = "na.or.complete")
}
df_out <- data.frame(country = unique(df$country_region))
df_out$YF_cor <- sapply(df_out$country, function(x) get_ma_corr(df, x))
df_out$BCG_cor <- sapply(df_out$country, function(x) get_ma_corr(df, x, "BCG"))
# get a nice figure
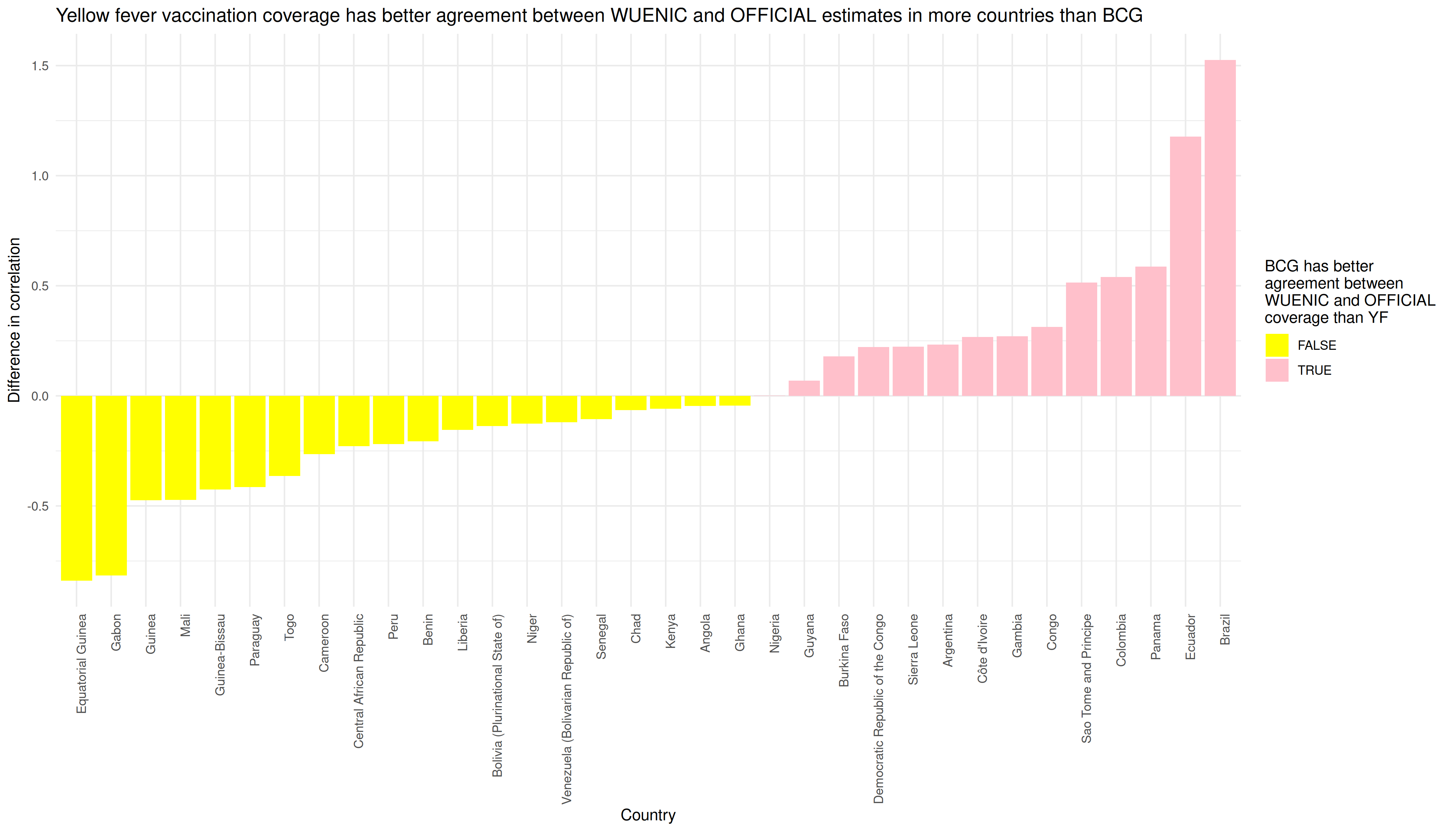
p <- df_out %>%
mutate(cor_diff = as.numeric(BCG_cor) - as.numeric(YF_cor)) %>%
filter(!is.na(cor_diff)) %>%
mutate(pos_neg = cor_diff > 0) %>%
ggplot() +
aes(x = reorder(country, cor_diff), y = cor_diff, fill = pos_neg) +
geom_col() +
theme_minimal() +
theme(axis.text.x = element_text(angle = 90, hjust = 1)) +
labs(x = "Country", y = "Difference in correlation",
fill = "BCG has better \nagreement between \nWUENIC and OFFICIAL \ncoverage than YF") +
scale_fill_manual(values = c("yellow", "pink")) +
ggtitle("Yellow fever vaccination coverage has better agreement between WUENIC and OFFICIAL estimates in more countries than BCG")
# save everything
ggsave(plot = p, filename = "BCG_YF_correlation_comparison.png",
width = 14, height = 8)
saveRDS(df, "wuenic.rds")
saveRDS(df_out, "corr_out.rds")Running this
id <- orderly_run("wuenic", echo = FALSE)
## Warning: Can't check if files are correctly gitignored
## ℹ Your outpack repo is in a subdirectory ''workdir/part2'' of your git repo
## ℹ To disable this check, set the option 'orderly_git_error_ignore' to TRUE by
## running `options(orderly_git_error_ignore = TRUE)`
## ℹ Starting packet 'wuenic' `20251007-071624-1130f836` at 2025-10-07 07:16:24.072161
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
##
## Attaching package: 'janitor'
## The following objects are masked from 'package:stats':
##
## chisq.test, fisher.test
## Warning in cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", : the
## standard deviation is zero
## Warning in cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", : the
## standard deviation is zero
## Warning in cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", : the
## standard deviation is zero
## Warning in cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", : the
## standard deviation is zero
## Warning in cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", : the
## standard deviation is zero
## Warning in cor(t(df_subset[df_subset$category %in% "WUENIC", grep("^x", : the
## standard deviation is zero
## ✔ Finished running 'wuenic.R'
## ! 6 warnings found:
## • the standard deviation is zero
## • the standard deviation is zero
## • the standard deviation is zero
## • the standard deviation is zero
## • the standard deviation is zero
## • the standard deviation is zero
## ℹ Finished 20251007-071624-1130f836 at 2025-10-07 07:16:26.292974 (2.220813 secs)The result
In graphical form
Documentation
Remaining questions
- Where do we put our starting data?
- How do we use Katy’s processed data
wuenic.rds? - What about custom packages?
- What about custom reusable code snippets, data and metadata?
Next steps
- On to collaboration
- Back to the project root
- Back to the geting started