Orderly Tutorial
What is orderly?
- A reproducible reporting framework
- A way of keeping track of versions of data
- A way of collaborating with other people
- A way of thinking about analysis
Original aims:
“With orderly we have two main hopes:
- analysts can write code that will straightforwardly run on someone else’s machine (or a remote machine)
- when an analysis that is run several times starts behaving differently it will be easy to see when the outputs started changing, and what inputs started changing at the same time”
(~ 2018)
But what is it?
- A package designed to make analysis more reproducible
- A way of structuring your analysis so to make it easy to understand, run and reuse
- A set of tools that make it easy to:
- track all inputs into an analysis (packages, code, and data resources)
- store multiple versions of an analysis where it is repeated
- track outputs of an analysis
- create analyses that depend on the outputs of previous analyses
Who uses it?
- Developed since May 2017 for the Vaccine Impact Modelling Consortium
- Used in the 2018-2020 DRC Ebola responses
- Used in the COVID-19 response, especially within the “real time modelling” group
- Used within research groups (HIV, Malaria, possibly others?)
Historical notes
orderly(version 1)- Created for Vaccine Impact Modelling Consortium and strongly focussed on reproducible research
- Used YAML everywhere
- Supported simple ways of working for a small centralised team
orderly2(soon to beorderly2.0.0)- A complete rewrite taking the best ideas from version 1 and dropping many less useful bits
- Easier to program against
- No more YAML
- Focusses on distributed collaborative analysis
- Also available as a python package!
Part 1: Getting started
Install orderly2
From the mrc-ide r-universe (recommended)
install.packages(
"orderly2",
repos = c("https://mrc-ide.r-universe.dev",
"https://cloud.r-project.org"))From GitHub using remotes:
From PyPi (for Python)
My first orderly report / task
There is a discussion to have here about naming. We might have this in a break…
The setup
First, load the package and create a new empty orderly root.
(for the rest of this section, we have setwd() into this directory; you should create an RStudio “Project” here.)
What’s in the box?
Create an empty report
Hello orderly world
We have edited src/example/example.R to contain:
Now we run
id <- orderly_run("example")
## ℹ Starting packet 'example' `20241023-074719-5042592d` at 2024-10-23 07:47:19.325677
## > d <- data.frame(greeting = "hello", to = "world")
## > write.csv(d, "hello.csv", row.names = FALSE)
## ✔ Finished running 'example.R'
## ℹ Finished 20241023-074719-5042592d at 2024-10-23 07:47:19.377219 (0.05154157 secs)Files created
- Directory named after the id (in
archive/example) - We have copied
example.Rinto the directory - Output sits next to inputs
- Metadata is stored in a hidden location
Contents of hello.csv:
Every packet has a unique id
and a bunch of metadata:
orderly_metadata(id)
## $schema_version
## [1] "0.1.1"
##
## $name
## [1] "example"
##
## $id
## [1] "20241023-074719-5042592d"
##
## $time
## $time$start
## [1] "2024-10-23 07:47:19 UTC"
##
## $time$end
## [1] "2024-10-23 07:47:19 UTC"
##
##
## $parameters
## NULL
##
## $files
## path size
## 1 example.R 95
## 2 hello.csv 32
## hash
## 1 sha256:541682d8b8dba9b2ddb4ac5809c03e6bedd58b52ab3e64a662f3f48e66a9639f
## 2 sha256:b9f0704f459f7ad9785ddee01a281d81f95a461dbb682436a263e0b7252e92b7
##
## $depends
## [1] packet query files
## <0 rows> (or 0-length row.names)
##
## $git
## $git$sha
## [1] "c6f469f362fd925206fad90a6749d5f9beaa6f02"
##
## $git$branch
## [1] "main"
##
## $git$url
## [1] "https://github.com/mrc-ide/orderly-tutorial"
##
##
## $custom
## $custom$orderly
## $custom$orderly$artefacts
## [1] description paths
## <0 rows> (or 0-length row.names)
##
## $custom$orderly$role
## path role
## 1 example.R orderly
##
## $custom$orderly$description
## $custom$orderly$description$display
## NULL
##
## $custom$orderly$description$long
## NULL
##
## $custom$orderly$description$custom
## NULL
##
##
## $custom$orderly$shared
## [1] here there
## <0 rows> (or 0-length row.names)
##
## $custom$orderly$session
## $custom$orderly$session$platform
## $custom$orderly$session$platform$version
## [1] "R version 4.4.1 (2024-06-14)"
##
## $custom$orderly$session$platform$os
## [1] "Ubuntu 22.04.5 LTS"
##
## $custom$orderly$session$platform$system
## [1] "x86_64, linux-gnu"
##
##
## $custom$orderly$session$packages
## package version attached
## 1 orderly2 1.99.48 TRUE
## 2 vctrs 0.6.5 FALSE
## 3 cli 3.6.3 FALSE
## 4 knitr 1.48 FALSE
## 5 rlang 1.1.4 FALSE
## 6 xfun 0.48 FALSE
## 7 jsonlite 1.8.9 FALSE
## 8 glue 1.8.0 FALSE
## 9 openssl 2.2.2 FALSE
## 10 askpass 1.2.1 FALSE
## 11 htmltools 0.5.8.1 FALSE
## 12 sys 3.4.3 FALSE
## 13 fansi 1.0.6 FALSE
## 14 rmarkdown 2.28 FALSE
## 15 evaluate 1.0.1 FALSE
## 16 tibble 3.2.1 FALSE
## 17 fastmap 1.2.0 FALSE
## 18 yaml 2.3.10 FALSE
## 19 lifecycle 1.0.4 FALSE
## 20 compiler 4.4.1 FALSE
## 21 fs 1.6.4 FALSE
## 22 pkgconfig 2.0.3 FALSE
## 23 digest 0.6.37 FALSE
## 24 gert 2.1.4 FALSE
## 25 R6 2.5.1 FALSE
## 26 utf8 1.2.4 FALSE
## 27 pillar 1.9.0 FALSE
## 28 credentials 2.0.2 FALSE
## 29 magrittr 2.0.3 FALSE
## 30 withr 3.0.1 FALSE
## 31 tools 4.4.1 FALSEWhat is a hash?
A one-way transformation from data to a fairly short string
Very small changes to the string give large changes to the hash
This means we can compare hashes and be confident we are looking at the same file (git does a lot of this)
Run it again, Sam
orderly_run("example")
## ℹ Starting packet 'example' `20241023-074719-752c6978` at 2024-10-23 07:47:19.462132
## > d <- data.frame(greeting = "hello", to = "world")
## > write.csv(d, "hello.csv", row.names = FALSE)
## ✔ Finished running 'example.R'
## ℹ Finished 20241023-074719-752c6978 at 2024-10-23 07:47:19.494556 (0.03242469 secs)
## [1] "20241023-074719-752c6978"- We have a new id with the new copy
A copy saved every time we run
Stop naming files data_final-rgf (2).csv, please
fs::dir_tree("workdir/part1")
## workdir/part1
## ├── archive
## │ └── example
## │ ├── 20241023-074719-5042592d
## │ │ ├── example.R
## │ │ └── hello.csv
## │ └── 20241023-074719-752c6978
## │ ├── example.R
## │ └── hello.csv
## ├── draft
## │ └── example
## ├── orderly_config.yml
## └── src
## └── example
## └── example.RA high-level overview of of packets:
Part 2: orderly code
What can I do?
- orderly code is any code you can use from R
- Use (almost) any package, any sort of file
But…
- The directory above your file does not exist
- Don’t use absolute paths or
../path fragments - You can add metadata to help future you/others
A clean beginning
Suppose we’re working on a data analysis pipeline, starting with “incoming data”:
Our setup:
Incoming data
I’ve copied some data in as data.xlsx into src/incoming.
- Modify
incoming.Rto tidy that up for consumption using your favourite packages. - Set your working directory to
src/incomingand just edit things as usual - Which sheet contains the data?
- Where is the data in that sheet?
- Do you like those column names?
- How about that date format?
Incoming data, cleaned
My attempt at cleaning:
d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
names(d) <- gsub(" ", "_", tolower(names(d)))
d$date <- as.Date(d$date)
write.csv(d, "data.csv", row.names = FALSE)- Read in the data
- Clean up the names (could have used
janitor) - Convert date format
- Write as
csv
Incoming data, running things
id <- orderly_run("incoming")
## ℹ Starting packet 'incoming' `20241023-074719-a46c26cc` at 2024-10-23 07:47:19.646878
## > d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
## > names(d) <- gsub(" ", "_", tolower(names(d)))
## > d$date <- as.Date(d$date)
## > write.csv(d, "data.csv", row.names = FALSE)
## ✔ Finished running 'incoming.R'
## ℹ Finished 20241023-074719-a46c26cc at 2024-10-23 07:47:19.678587 (0.03170943 secs)Our generated metadata:
orderly_metadata(id)
## $schema_version
## [1] "0.1.1"
##
## $name
## [1] "incoming"
##
## $id
## [1] "20241023-074719-a46c26cc"
##
## $time
## $time$start
## [1] "2024-10-23 07:47:19 UTC"
##
## $time$end
## [1] "2024-10-23 07:47:19 UTC"
##
##
## $parameters
## NULL
##
## $files
## path size
## 1 data.csv 466
## 2 data.xlsx 10851
## 3 incoming.R 174
## hash
## 1 sha256:9117700f079b786812cc20904f4c34f5659f2986a923d2771216551cf378e86f
## 2 sha256:149445ecc545eb987ecc0d5255a48f21165ee1c9b6b54c84b882ce1fbda066b7
## 3 sha256:8ce9dc59614d62b39cd4ff8cfba08e48f993a9be6d3c29357118df386bf7688f
##
## $depends
## [1] packet query files
## <0 rows> (or 0-length row.names)
##
## $git
## $git$sha
## [1] "c6f469f362fd925206fad90a6749d5f9beaa6f02"
##
## $git$branch
## [1] "main"
##
## $git$url
## [1] "https://github.com/mrc-ide/orderly-tutorial"
##
##
## $custom
## $custom$orderly
## $custom$orderly$artefacts
## [1] description paths
## <0 rows> (or 0-length row.names)
##
## $custom$orderly$role
## path role
## 1 incoming.R orderly
##
## $custom$orderly$description
## $custom$orderly$description$display
## NULL
##
## $custom$orderly$description$long
## NULL
##
## $custom$orderly$description$custom
## NULL
##
##
## $custom$orderly$shared
## [1] here there
## <0 rows> (or 0-length row.names)
##
## $custom$orderly$session
## $custom$orderly$session$platform
## $custom$orderly$session$platform$version
## [1] "R version 4.4.1 (2024-06-14)"
##
## $custom$orderly$session$platform$os
## [1] "Ubuntu 22.04.5 LTS"
##
## $custom$orderly$session$platform$system
## [1] "x86_64, linux-gnu"
##
##
## $custom$orderly$session$packages
## package version attached
## 1 orderly2 1.99.48 TRUE
## 2 vctrs 0.6.5 FALSE
## 3 cli 3.6.3 FALSE
## 4 knitr 1.48 FALSE
## 5 rlang 1.1.4 FALSE
## 6 xfun 0.48 FALSE
## 7 jsonlite 1.8.9 FALSE
## 8 glue 1.8.0 FALSE
## 9 openssl 2.2.2 FALSE
## 10 askpass 1.2.1 FALSE
## 11 htmltools 0.5.8.1 FALSE
## 12 sys 3.4.3 FALSE
## 13 readxl 1.4.3 FALSE
## 14 fansi 1.0.6 FALSE
## 15 rmarkdown 2.28 FALSE
## 16 cellranger 1.1.0 FALSE
## 17 evaluate 1.0.1 FALSE
## 18 tibble 3.2.1 FALSE
## 19 fastmap 1.2.0 FALSE
## 20 yaml 2.3.10 FALSE
## 21 lifecycle 1.0.4 FALSE
## 22 compiler 4.4.1 FALSE
## 23 fs 1.6.4 FALSE
## 24 pkgconfig 2.0.3 FALSE
## 25 digest 0.6.37 FALSE
## 26 gert 2.1.4 FALSE
## 27 R6 2.5.1 FALSE
## 28 utf8 1.2.4 FALSE
## 29 pillar 1.9.0 FALSE
## 30 credentials 2.0.2 FALSE
## 31 magrittr 2.0.3 FALSE
## 32 withr 3.0.1 FALSE
## 33 tools 4.4.1 FALSE“Resources”
- Any file that is an input
- For example:
- Scripts that you
source() - R Markdown files for
knitrorrmarkdown - Data files (
.csv,.xlsx, etc) - Plain text files (
README.md, licence info, etc)
- Scripts that you
- Here,
data.xlsxis an input
Telling orderly about resources
orderly_resource("data.xlsx")
d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
names(d) <- gsub(" ", "_", tolower(names(d)))
d$date <- as.Date(d$date)
write.csv(d, "data.csv", row.names = FALSE)- Tells orderly
data.xlsxis a resource - Fail early if resource not found
- Error if resource is modified
- Extra metadata, advertising what files were used
id <- orderly_run("incoming")
## ℹ Starting packet 'incoming' `20241023-074719-bbb08e35` at 2024-10-23 07:47:19.737755
## > orderly_resource("data.xlsx")
## > d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
## > names(d) <- gsub(" ", "_", tolower(names(d)))
## > d$date <- as.Date(d$date)
## > write.csv(d, "data.csv", row.names = FALSE)
## ✔ Finished running 'incoming.R'
## ℹ Finished 20241023-074719-bbb08e35 at 2024-10-23 07:47:19.764167 (0.02641225 secs)
orderly_metadata(id)$custom$orderly$role
## path role
## 1 incoming.R orderly
## 2 data.xlsx resource“Artefacts”
- Any file that is an output
- For example:
- Datasets you generate
- html or pdf output from
knitrorrmarkdown - Plain text files
- Inputs themselves, sometimes
- Here,
data.csvis an artefact
Telling orderly about artefacts
orderly_resource("data.xlsx")
orderly_artefact(files = "data.csv", description = "Cleaned data")
d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
names(d) <- gsub(" ", "_", tolower(names(d)))
d$date <- as.Date(d$date)
write.csv(d, "data.csv", row.names = FALSE)- Tells orderly
csv.xlsxis an artefact - Fail if artefact not produced
- Extra metadata, advertising what files were produced
id <- orderly_run("incoming")
## ℹ Starting packet 'incoming' `20241023-074719-cf0bc2a0` at 2024-10-23 07:47:19.813293
## > orderly_resource("data.xlsx")
## > orderly_artefact(files = "data.csv", description = "Cleaned data")
## > d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
## > names(d) <- gsub(" ", "_", tolower(names(d)))
## > d$date <- as.Date(d$date)
## > write.csv(d, "data.csv", row.names = FALSE)
## ✔ Finished running 'incoming.R'
## ℹ Finished 20241023-074719-cf0bc2a0 at 2024-10-23 07:47:19.844239 (0.03094578 secs)
orderly_metadata(id)$custom$orderly$artefacts
## description paths
## 1 Cleaned data data.csvMore metadata
orderly_description(
display = "Incoming data from Otherlandia",
long = "Data as given to us from the MoH in Otherlandia.",
custom = list(received = "2024-10-22"))
orderly_resource("data.xlsx")
orderly_artefact(files = "data.csv", description = "Cleaned data")
d <- readxl::read_excel("data.xlsx", sheet = 2, skip = 2)
names(d) <- gsub(" ", "_", tolower(names(d)))
d$date <- as.Date(d$date)
write.csv(d, "data.csv", row.names = FALSE)Running this:
id <- orderly_run("incoming", echo = FALSE)
## ℹ Starting packet 'incoming' `20241023-074719-e393a4e3` at 2024-10-23 07:47:19.893583
## ✔ Finished running 'incoming.R'
## ℹ Finished 20241023-074719-e393a4e3 at 2024-10-23 07:47:19.920295 (0.02671218 secs)
orderly_metadata(id)$custom$orderly$description
## $display
## [1] "Incoming data from Otherlandia"
##
## $long
## [1] "Data as given to us from the MoH in Otherlandia."
##
## $custom
## $custom$received
## [1] "2024-10-22"Dependencies
- This is really the point of orderly
- You can pull in any file from any previously run packet
- You can use queries to select packets to depend on
Our aim: We want to use data.csv in some analysis
Setting up a dependency
orderly_dependency("incoming", "latest", "data.csv")
orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
description = "Plots of coverage")
d <- read.csv("data.csv")
d$date <- as.Date(d$date)
png("coverage-gf.png")
plot(gf_coverage ~ date, d, type = "l")
dev.off()
png("coverage-bf.png")
plot(bf_coverage ~ date, d, type = "l")
dev.off()This is the only file within our analysis directory:
Running the report
id <- orderly_run("analysis")
## ℹ Starting packet 'analysis' `20241023-074719-fca3dc27` at 2024-10-23 07:47:19.991456
## > orderly_dependency("incoming", "latest", "data.csv")
## ℹ Depending on incoming @ `20241023-074719-e393a4e3` (via latest(name == "incoming"))
## > orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
## + description = "Plots of coverage")
## > d <- read.csv("data.csv")
## > d$date <- as.Date(d$date)
## > png("coverage-gf.png")
## > plot(gf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## > png("coverage-bf.png")
## > plot(bf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## ✔ Finished running 'analysis.R'
## ℹ Finished 20241023-074719-fca3dc27 at 2024-10-23 07:47:20.083316 (0.09186029 secs)The aftermath
fs::dir_tree("workdir/part2")
## workdir/part2
## ├── archive
## │ ├── analysis
## │ │ └── 20241023-074719-fca3dc27
## │ │ ├── analysis.R
## │ │ ├── coverage-bf.png
## │ │ ├── coverage-gf.png
## │ │ └── data.csv
## │ └── incoming
## │ ├── 20241023-074719-a46c26cc
## │ │ ├── data.csv
## │ │ ├── data.xlsx
## │ │ └── incoming.R
## │ ├── 20241023-074719-bbb08e35
## │ │ ├── data.csv
## │ │ ├── data.xlsx
## │ │ └── incoming.R
## │ ├── 20241023-074719-cf0bc2a0
## │ │ ├── data.csv
## │ │ ├── data.xlsx
## │ │ └── incoming.R
## │ └── 20241023-074719-e393a4e3
## │ ├── data.csv
## │ ├── data.xlsx
## │ └── incoming.R
## ├── draft
## │ ├── analysis
## │ └── incoming
## ├── orderly_config.yml
## └── src
## ├── analysis
## │ └── analysis.R
## └── incoming
## ├── data.xlsx
## └── incoming.RSome comments on this
- The
data.csvfile has been copied from the final copy ofincomingintoanalysis - The dependency system works interactively too (try it!)
- The logs indicate how dependency resolution occurred
- Metadata about the dependencies is included:
Using specific versions
orderly_dependency("incoming", "20241023-074719-bbb08e35", "data.csv")
orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
description = "Plots of coverage")
d <- read.csv("data.csv")
d$date <- as.Date(d$date)
png("coverage-gf.png")
plot(gf_coverage ~ date, d, type = "l")
dev.off()
png("coverage-bf.png")
plot(bf_coverage ~ date, d, type = "l")
dev.off()Running this
id <- orderly_run("analysis")
## ℹ Starting packet 'analysis' `20241023-074720-27b9baf4` at 2024-10-23 07:47:20.159643
## > orderly_dependency("incoming", "20241023-074719-bbb08e35", "data.csv")
## ℹ Depending on incoming @ `20241023-074719-bbb08e35` (via single(id == "20241023-074719-bbb08e35" && name == "incoming"))
## > orderly_artefact(files = c("coverage-gf.png", "coverage-bf.png"),
## + description = "Plots of coverage")
## > d <- read.csv("data.csv")
## > d$date <- as.Date(d$date)
## > png("coverage-gf.png")
## > plot(gf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## > png("coverage-bf.png")
## > plot(bf_coverage ~ date, d, type = "l")
## > dev.off()
## png
## 2
## ✔ Finished running 'analysis.R'
## ℹ Finished 20241023-074720-27b9baf4 at 2024-10-23 07:47:20.215387 (0.05574393 secs)with metadata
Part 3: Collaboration
Working with other people
- Where do you store your code?
- Where do you store your data?
- Where do you store your outputs?
- How will things change over time?
- Is it sensitive?
The setup
Here we ignore the git side for now and focus on sharing outputs
Thom Rawson has kindly set up a bunch of case data for us to use. He’s put it in an orderly root that we can use as an orderly location.
The sausage factory
We’ll hide this in the final build
local({
path_upstream <- "workdir/part3-upstream"
orderly_init(path_upstream)
orderly_init(path_upstream)
orderly_new("cases", root = path_upstream)
fs::file_copy("inputs/part3/cases.R",
file.path(path_upstream, "src/cases/cases.R"),
overwrite = TRUE)
re <- "^(.+)-2020\\.csv$"
files <- dir("inputs/part3/cases", re)
regions <- sub(re, "\\1", files)
names(files) <- regions
for (region in regions) {
fs::file_copy(file.path("inputs/part3/cases", files[[region]]),
file.path(path_upstream, "src/cases/cases.csv"),
overwrite = TRUE)
orderly_run("cases", list(region = region, year = 2020), root = path_upstream)
}
})
## ✔ Created orderly root at '/home/runner/work/orderly-tutorial/orderly-tutorial/workdir/part3-upstream'
## ✔ Wrote '.gitignore'
## ✔ Created 'src/cases/cases.R'
## ℹ Starting packet 'cases' `20241023-074720-562a1556` at 2024-10-23 07:47:20.347022
## ℹ Parameters:
## • region: east_of_england
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-562a1556 at 2024-10-23 07:47:20.38153 (0.03450727 secs)
## ℹ Starting packet 'cases' `20241023-074720-67c51964` at 2024-10-23 07:47:20.411574
## ℹ Parameters:
## • region: london
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-67c51964 at 2024-10-23 07:47:20.443204 (0.03163028 secs)
## ℹ Starting packet 'cases' `20241023-074720-79050123` at 2024-10-23 07:47:20.479728
## ℹ Parameters:
## • region: midlands
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-79050123 at 2024-10-23 07:47:20.511881 (0.03215313 secs)
## ℹ Starting packet 'cases' `20241023-074720-89dc11be` at 2024-10-23 07:47:20.544771
## ℹ Parameters:
## • region: north_east_and_yorkshire
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-89dc11be at 2024-10-23 07:47:20.580047 (0.0352757 secs)
## ℹ Starting packet 'cases' `20241023-074720-9af20659` at 2024-10-23 07:47:20.611648
## ℹ Parameters:
## • region: north_west
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-9af20659 at 2024-10-23 07:47:20.643338 (0.03169012 secs)
## ℹ Starting packet 'cases' `20241023-074720-ab86950a` at 2024-10-23 07:47:20.676958
## ℹ Parameters:
## • region: south_east
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-ab86950a at 2024-10-23 07:47:20.715482 (0.03852415 secs)
## ℹ Starting packet 'cases' `20241023-074720-bde9b16f` at 2024-10-23 07:47:20.748124
## ℹ Parameters:
## • region: south_west
## • year: 2020
## > orderly_parameters(region = NULL, year = NULL)
## > orderly_resource(files = "cases.csv")
## > orderly_artefact(files = "cases.csv", description = "Case data")
## ✔ Finished running 'cases.R'
## ℹ Finished 20241023-074720-bde9b16f at 2024-10-23 07:47:20.778946 (0.03082228 secs)Adding a location
- Here we have used the
pathlocation type - We will try this with the
packittype in the workshop - Locations are just another orderly root where you can find packets
- We use
fs::path_abs()for now, for uninteresting reasons
What has Thom been up to?
orderly_location_pull_metadata()
## ℹ Fetching metadata from 1 location: 'thom'
## ✔ Found 7 packets at 'thom', of which 7 are new
orderly_metadata_extract(location = "thom")
## id name parameters
## 1 20241023-074720-562a1556 cases east_of_....
## 2 20241023-074720-67c51964 cases london, 2020
## 3 20241023-074720-79050123 cases midlands....
## 4 20241023-074720-89dc11be cases north_ea....
## 5 20241023-074720-9af20659 cases north_we....
## 6 20241023-074720-ab86950a cases south_ea....
## 7 20241023-074720-bde9b16f cases south_we....Slightly easier to read, but harder to write
orderly_metadata_extract(
location = "thom",
extract = c("name",
region = "parameters.region is string",
year = "parameters.year is number"))
## id name region year
## 1 20241023-074720-562a1556 cases east_of_england 2020
## 2 20241023-074720-67c51964 cases london 2020
## 3 20241023-074720-79050123 cases midlands 2020
## 4 20241023-074720-89dc11be cases north_east_and_yorkshire 2020
## 5 20241023-074720-9af20659 cases north_west 2020
## 6 20241023-074720-ab86950a cases south_east 2020
## 7 20241023-074720-bde9b16f cases south_west 2020Depending on this
And code:
Query syntax
orderly_search("latest", name = "cases", location = "thom")
## [1] "20241023-074720-bde9b16f"
orderly_search("latest(parameter:region == 'east_of_england')", name = "cases",
location = "thom")
## [1] "20241023-074720-562a1556"
orderly_search(
"latest(parameter:region == 'london' && parameter:year == 2020)",
name = "cases",
location = "thom")
## [1] "20241023-074720-67c51964"You can use this elsewhere:
The files argument
“Save the file cases.csv as london.csv in the working version”
Run the new report
orderly_run("analysis")
## ℹ Starting packet 'analysis' `20241023-074720-f4fe1ce6` at 2024-10-23 07:47:20.961586
## > orderly_dependency("cases",
## + 'latest(parameter:region == "london")',
## + c("london.csv" = "cases.csv"))
## ✖ Error running 'analysis.R'
## ℹ Finished 20241023-074720-f4fe1ce6 at 2024-10-23 07:47:21.005248 (0.04366183 secs)
## Error in `orderly_run()`:
## ! Failed to run report
## Caused by error in `outpack_packet_use_dependency()`:
## ! Failed to find packet for query 'latest(parameter:region == "london"
## && name == "cases")'
## ℹ See 'rlang::last_error()$explanation' for detailsoh no
Using packets from elsewhere
Two options
- Pull the packet to make it local (
orderly_location_pull_packet) - Tell orderly which locations to use
Running with location
id <- orderly_run("analysis", location = "thom") # or allow_remote = TRUE
## ℹ Starting packet 'analysis' `20241023-074721-0f28013a` at 2024-10-23 07:47:21.063695
## > orderly_dependency("cases",
## + 'latest(parameter:region == "london")',
## + c("london.csv" = "cases.csv"))
## ℹ Looking for suitable files already on disk
## ℹ Need to fetch 1 file (669.9 kB) from 1 location
## ℹ Depending on cases @ `20241023-074720-67c51964` (via latest(parameter:region == "london" && name == "cases"))
## > d <- read.csv("london.csv")
## > png("london.png")
## > plot(Week_Cases ~ Week, d)
## > dev.off()
## png
## 2
## ✔ Finished running 'analysis.R'
## ℹ Finished 20241023-074721-0f28013a at 2024-10-23 07:47:21.178957 (0.115262 secs)- Where dependencies are resolved is a property of
orderly_run(), not the source of the report - Files are fetched as we run - only used files are copied
The result
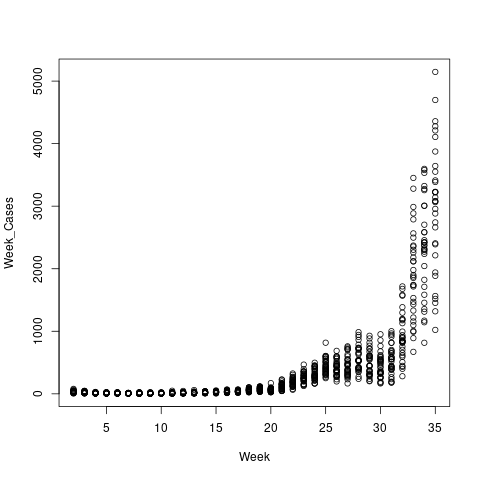
Include many dependencies
Thom has new data for us!
orderly_metadata_extract(
location = "thom",
extract = c("name",
region = "parameters.region is string",
year = "parameters.year is number"))
## id name region year
## 1 20241023-074720-562a1556 cases east_of_england 2020
## 2 20241023-074720-67c51964 cases london 2020
## 3 20241023-074720-79050123 cases midlands 2020
## 4 20241023-074720-89dc11be cases north_east_and_yorkshire 2020
## 5 20241023-074720-9af20659 cases north_west 2020
## 6 20241023-074720-ab86950a cases south_east 2020
## 7 20241023-074720-bde9b16f cases south_west 2020
## 8 20241023-074721-3ced1c27 cases london 2021Run our analysis with this data
This time we try pulling
orderly_location_pull_packet("latest", name = "cases", location = "thom")
## ℹ Looking for suitable files already on disk
## ✔ Found 1 file on disk
## ℹ Need to fetch 1 file (150 B) from 1 location
id <- orderly_run("analysis")
## ℹ Starting packet 'analysis' `20241023-074721-672ddcb4` at 2024-10-23 07:47:21.407465
## > orderly_dependency("cases",
## + 'latest(parameter:region == "london")',
## + c("london.csv" = "cases.csv"))
## ℹ Depending on cases @ `20241023-074721-3ced1c27` (via latest(parameter:region == "london" && name == "cases"))
## > d <- read.csv("london.csv")
## > png("london.png")
## > plot(Week_Cases ~ Week, d)
## > dev.off()
## png
## 2
## ✔ Finished running 'analysis.R'
## ℹ Finished 20241023-074721-672ddcb4 at 2024-10-23 07:47:21.491953 (0.0844872 secs)Some thoughts
Documentation
Details for writing reports/tasks
- Shared resources
- Resources
- Loops over dependencies
- Metadata
What is saved?
- Information about files you consumed, and produced
Ways of collaborating
Assorted issues
Misc
- How to get data into orderly in the first place
- git versioned files
- git ignored files
- files from canonical locations
- databases
- Coping with failure
- Running knitr/rmarkdown
- Getting files out of orderly
The right number of packets
- Similar to “how big is a git repo”
- Some issues:
- People fragmenting packets to overcome flakey analysis
- Millions of packets, leading to complex and slow queries
- Hard to get the right combination of packets
The right number of parameters
- Too few is too inflexible
- Too many
Interaction with git
- Don’t save outputs
- Save some inputs?
- Don’t save secrets
- Don’t save locations