An introduction to odin2 and dust2
odin
- A “domain specific language”
- Originally designed for ordinary differential equations
- Some very basic discrete time/stochastic support
odin
ODE models
\[\begin{gather*} \frac{dS}{dt} = -\beta S \frac{I}{N}\\ \frac{dI}{dt} = \beta S \frac{I}{N} - \gamma I\\ \frac{dR}{dt} = \gamma I \end{gather*}\]
Things to note:
- out of order definition
- every variable has
initialandderivpair
Compiling the model with odin2
Running the model with dust2
The output has dimensions number of states x number of timepoints
Unpacking states
Output can be nicely unpacked into the different states using dust_unpack_state
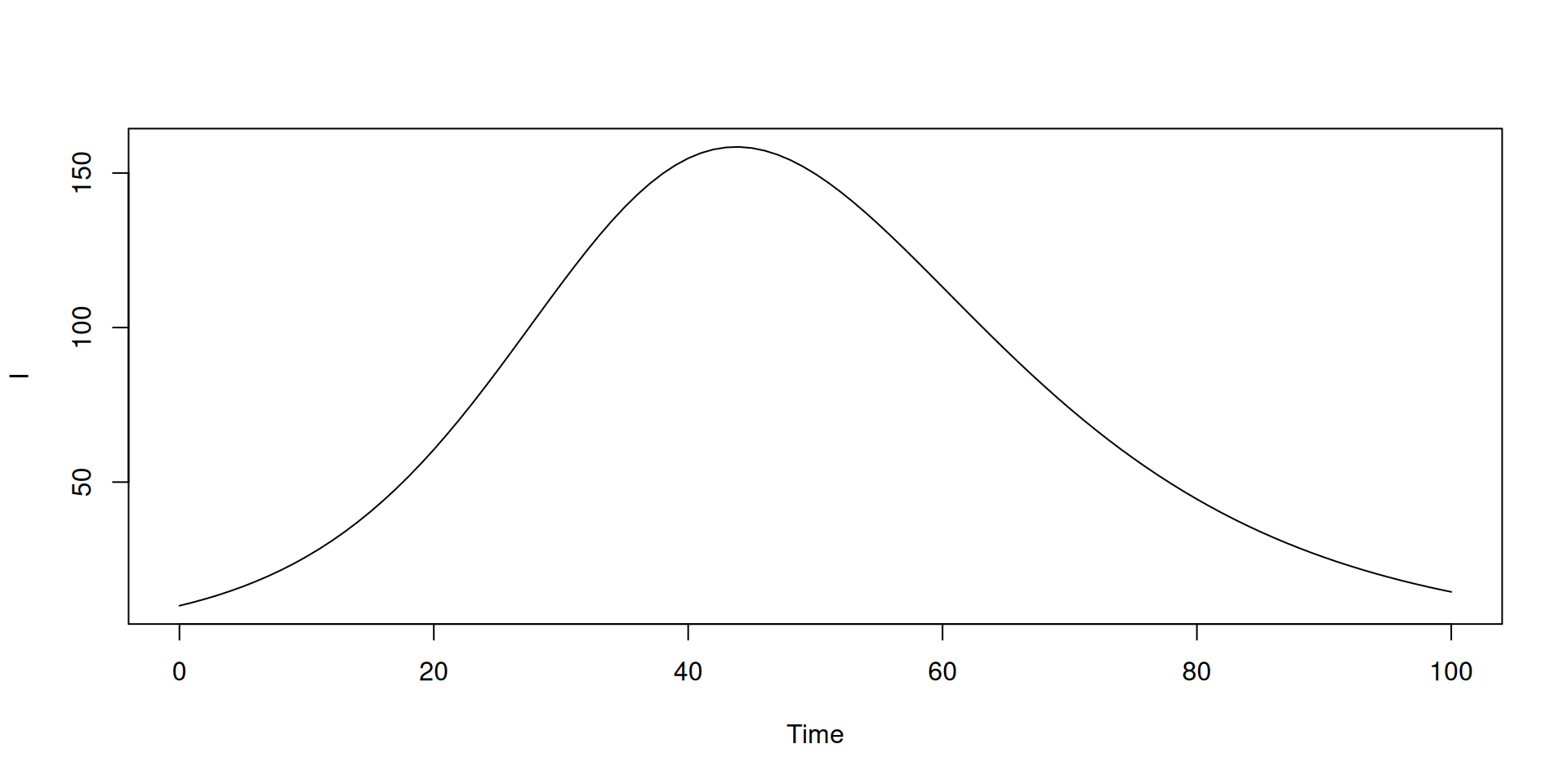
Stochastic models
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
initial(S) <- N - I0
initial(I) <- I0
initial(R) <- 0
p_SI <- 1 - exp(-beta * I / N * dt)
p_IR <- 1 - exp(-gamma * dt)
n_SI <- Binomial(S, p_SI)
n_IR <- Binomial(I, p_IR)
N <- parameter(1000)
I0 <- parameter(10)
beta <- parameter(0.2)
gamma <- parameter(0.1)\[\begin{gather*} S(t + \Delta t) = S(t) - n_{SI}\\ I(t + \Delta t) = I(t) + n_{SI} - n_{IR}\\ R(t + \Delta t) = R(t) + n_{IR} \end{gather*}\]
dtis a special parameter- every variable has
initialandupdatepair
…compared with ODE models
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
initial(S) <- N - I0
initial(I) <- I0
initial(R) <- 0
p_SI <- 1 - exp(-beta * I / N * dt)
p_IR <- 1 - exp(-gamma * dt)
n_SI <- Binomial(S, p_SI)
n_IR <- Binomial(I, p_IR)
N <- parameter(1000)
I0 <- parameter(10)
beta <- parameter(0.2)
gamma <- parameter(0.1)Compiling with odin2
sir <- odin({
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
initial(S) <- N - I0
initial(I) <- I0
initial(R) <- 0
p_SI <- 1 - exp(-beta * I / N * dt)
p_IR <- 1 - exp(-gamma * dt)
n_SI <- Binomial(S, p_SI)
n_IR <- Binomial(I, p_IR)
N <- parameter(1000)
I0 <- parameter(10)
beta <- parameter(0.2)
gamma <- parameter(0.1)
})Running a single simulation
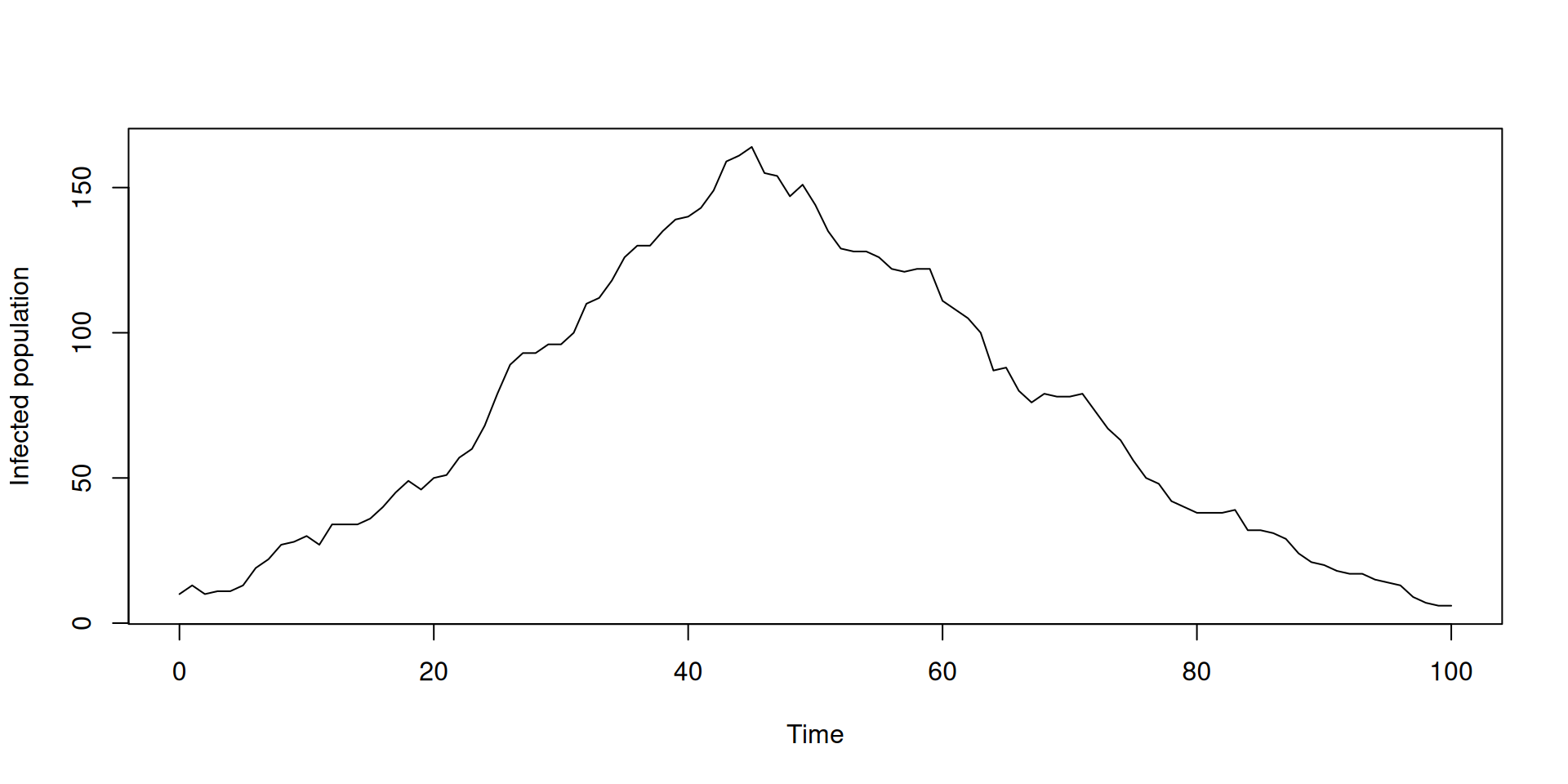
Running multiple simulations
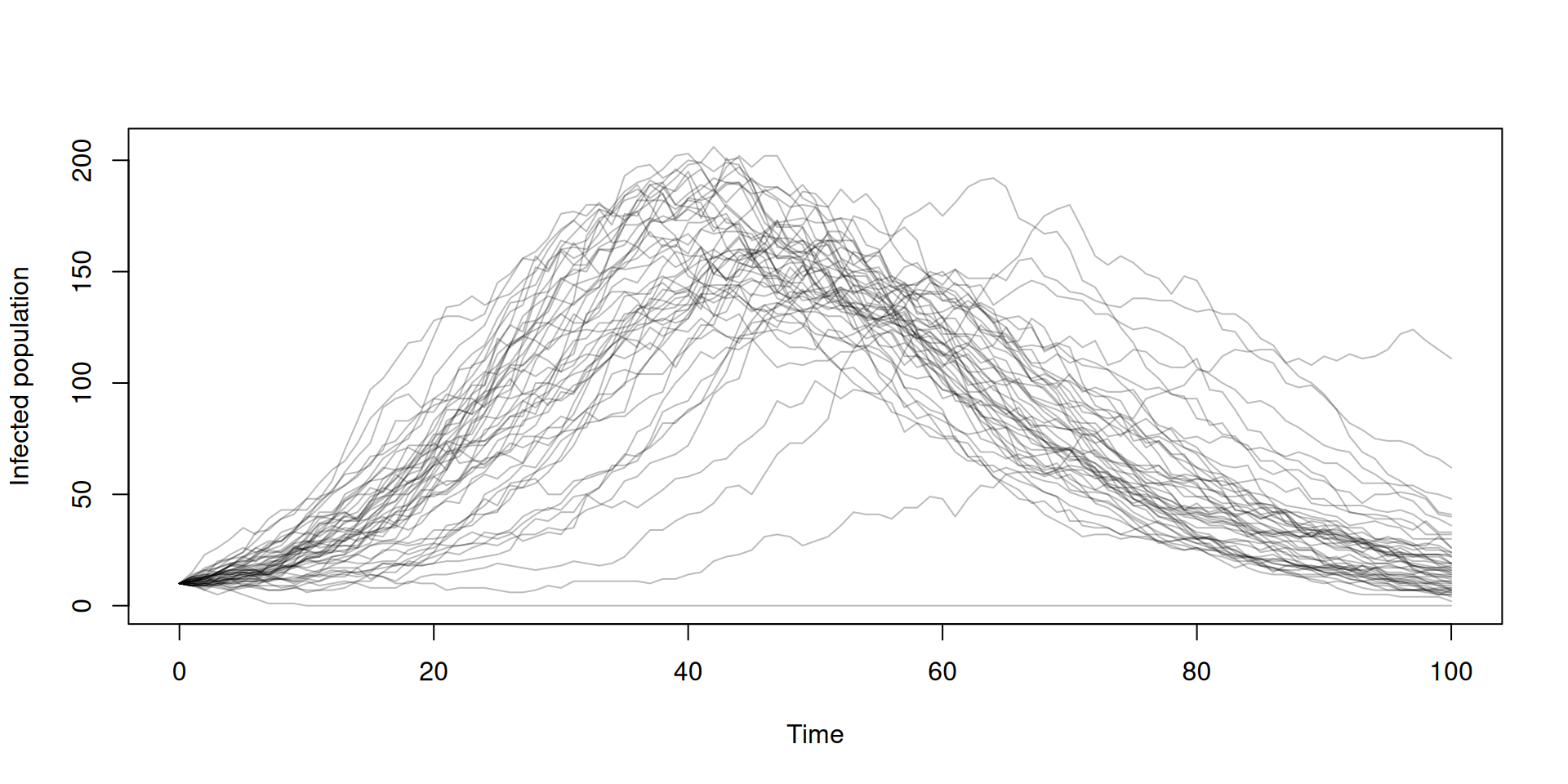
Calculating incidence with zero_every
sir <- odin({
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
update(incidence) <- incidence + n_SI
p_SI <- 1 - exp(-beta * I / N * dt)
p_IR <- 1 - exp(-gamma * dt)
n_SI <- Binomial(S, p_SI)
n_IR <- Binomial(I, p_IR)
initial(S) <- N - I0
initial(I) <- I0
initial(R) <- 0
initial(incidence, zero_every = 1) <- 0
N <- parameter(1000)
I0 <- parameter(10)
beta <- parameter(0.2)
gamma <- parameter(0.1)
})Incidence accumulates then resets
sys <- dust_system_create(sir, pars = list(), dt = 1 / 128)
dust_system_set_state_initial(sys)
t <- seq(0, 20, by = 1 / 128)
y <- dust_system_simulate(sys, t)
y <- dust_unpack_state(sys, y)
plot(t[t %% 1 == 0], y$incidence[t %% 1 == 0], type = "o", pch = 19,
ylab = "Infection incidence", xlab = "Time")
lines(t, y$incidence, col = "red")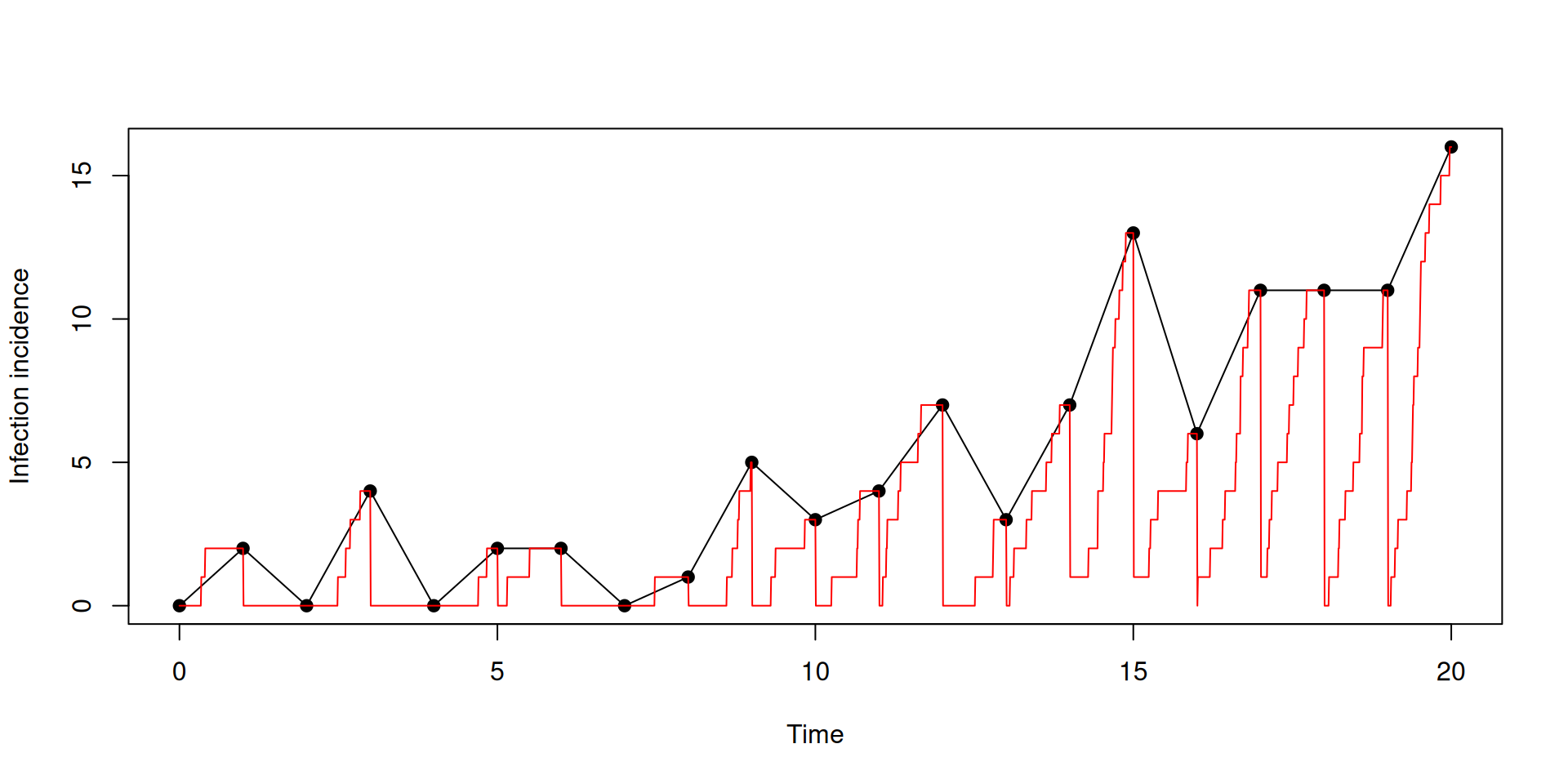
Time-varying inputs: using time
sir <- odin({
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
update(incidence) <- incidence + n_SI
seed <- if (time == seed_time) seed_size else 0
p_SI <- 1 - exp(-beta * I / N * dt)
p_IR <- 1 - exp(-gamma * dt)
n_SI <- min(seed + Binomial(S, p_SI), S)
n_IR <- Binomial(I, p_IR)
initial(S) <- N
initial(I) <- 0
initial(R) <- 0
initial(incidence, zero_every = 1) <- 0
N <- parameter(1000)
beta <- parameter(0.2)
gamma <- parameter(0.1)
seed_time <- parameter()
seed_size <- parameter()
})Time-varying inputs: using time
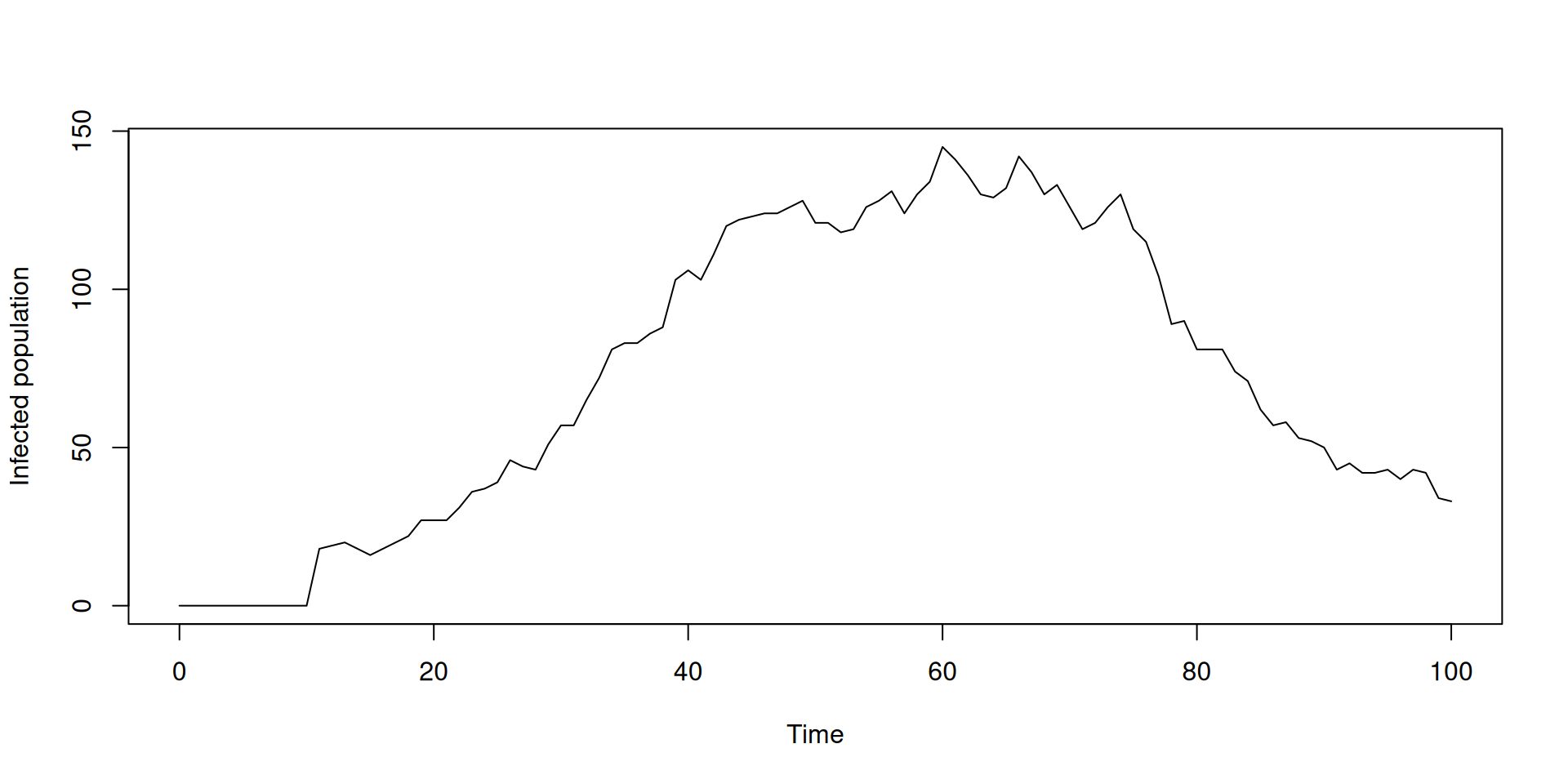
Time-varying inputs: using interpolate
The interpolate function in odin can be used for time-varying parameters, with specification of
- the times of changepoints
- the values at those changepoints
- the type of interpolation: linear, constant or spline
Time-varying inputs: using interpolate
sir <- odin({
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
update(incidence) <- incidence + n_SI
beta <- interpolate(beta_time, beta_value, "constant")
beta_time <- parameter()
beta_value <- parameter()
dim(beta_time, beta_value) <- parameter(rank = 1)
p_SI <- 1 - exp(-beta * I / N * dt)
p_IR <- 1 - exp(-gamma * dt)
n_SI <- Binomial(S, p_SI)
n_IR <- Binomial(I, p_IR)
initial(S) <- N - I0
initial(I) <- I0
initial(R) <- 0
initial(incidence, zero_every = 1) <- 0
N <- parameter(1000)
I0 <- parameter(10)
gamma <- parameter(0.1)
})Time-varying inputs: using interpolate
pars <- list(beta_time = c(0, 30), beta_value = c(0.2, 1))
sys <- dust_system_create(sir, pars = pars, dt = 0.25)
dust_system_set_state_initial(sys)
t <- seq(0, 100)
y <- dust_system_simulate(sys, t)
y <- dust_unpack_state(sys, y)
plot(t, y$I, type = "l", xlab = "Time", ylab = "Infected population")
abline(v = 30, lty = 3)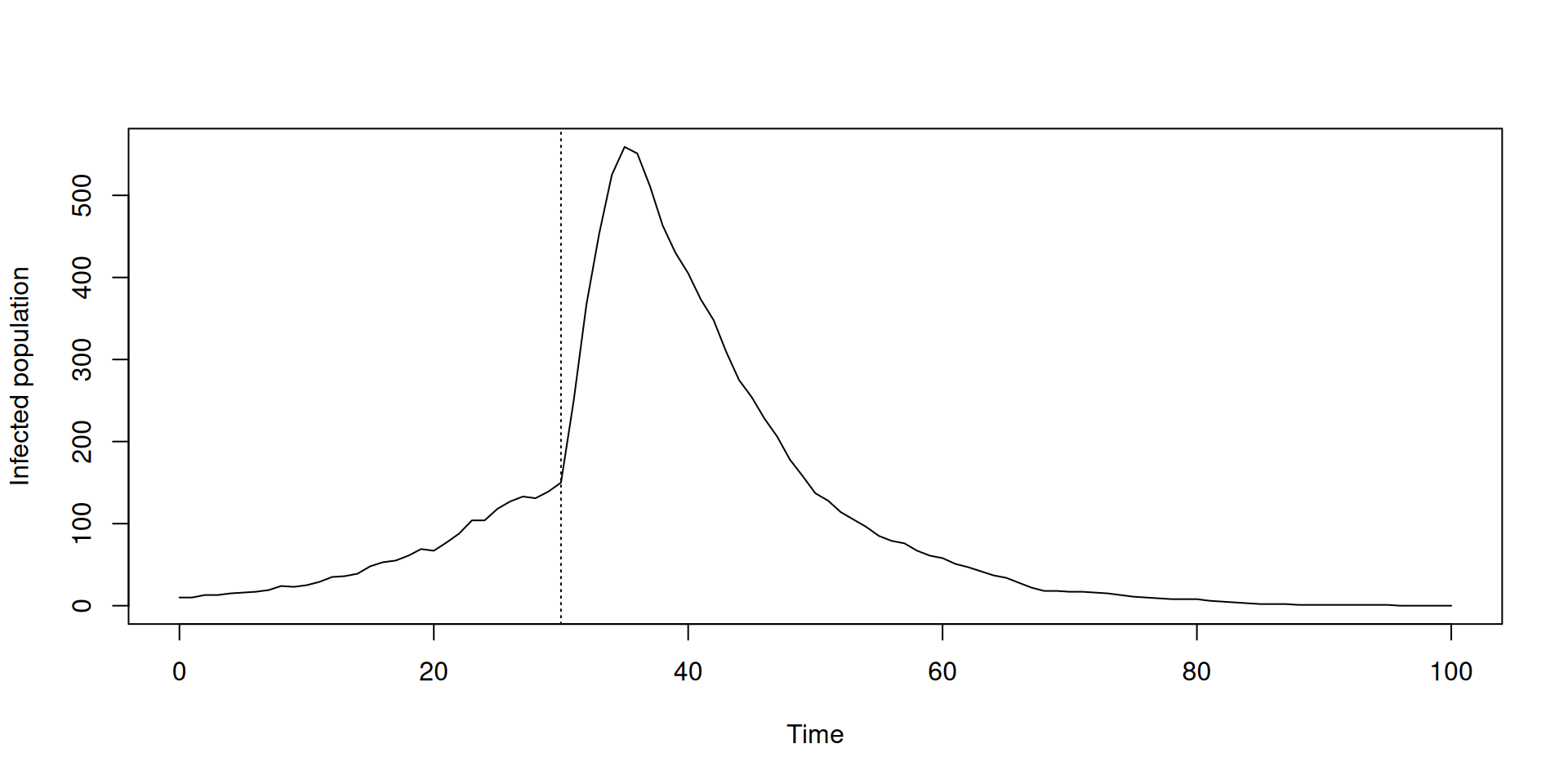
Using arrays
sir_age <- odin({
# Equations for transitions between compartments by age group
update(S[]) <- S[i] - n_SI[i]
update(I[]) <- I[i] + n_SI[i] - n_IR[i]
update(R[]) <- R[i] + n_IR[i]
update(incidence) <- incidence + sum(n_SI)
# Individual probabilities of transition:
p_SI[] <- 1 - exp(-lambda[i] * dt) # S to I
p_IR <- 1 - exp(-gamma * dt) # I to R
# Calculate force of infection
# age-structured contact matrix: m[i, j] is mean number
# of contacts an individual in group i has with an
# individual in group j per time unit
m <- parameter()
# here s_ij[i, j] gives the mean number of contacts an
# individual in group i will have with the currently
# infectious individuals of group j
s_ij[, ] <- m[i, j] * I[j]
# lambda[i] is the total force of infection on an
# individual in group i
lambda[] <- beta * sum(s_ij[i, ])
# Draws from binomial distributions for numbers
# changing between compartments:
n_SI[] <- Binomial(S[i], p_SI[i])
n_IR[] <- Binomial(I[i], p_IR)
initial(S[]) <- S0[i]
initial(I[]) <- I0[i]
initial(R[]) <- 0
initial(incidence, zero_every = 1) <- 0
S0 <- parameter()
I0 <- parameter()
beta <- parameter(0.2)
gamma <- parameter(0.1)
n_age <- parameter()
dim(S, S0, n_SI, p_SI) <- n_age
dim(I, I0, n_IR) <- n_age
dim(R) <- n_age
dim(m, s_ij) <- c(n_age, n_age)
dim(lambda) <- n_age
})Using arrays
Remember: m[i, j] is the mean number of contacts per time unit for an individual in group i has with an individual in group j
Using arrays
sys <- dust_system_create(sir_age, pars = pars, dt = 0.25)
dust_system_set_state_initial(sys)
t <- seq(0, 100)
y <- dust_system_simulate(sys, t)
y <- dust_unpack_state(sys, y)
matplot(t, t(y$I), type = "l", lty = 1, col = c("red", "blue"),
xlab = "Time", ylab = "Infected population")
legend("topright", c("children", "adults"), col = c("red", "blue"), lty = 1)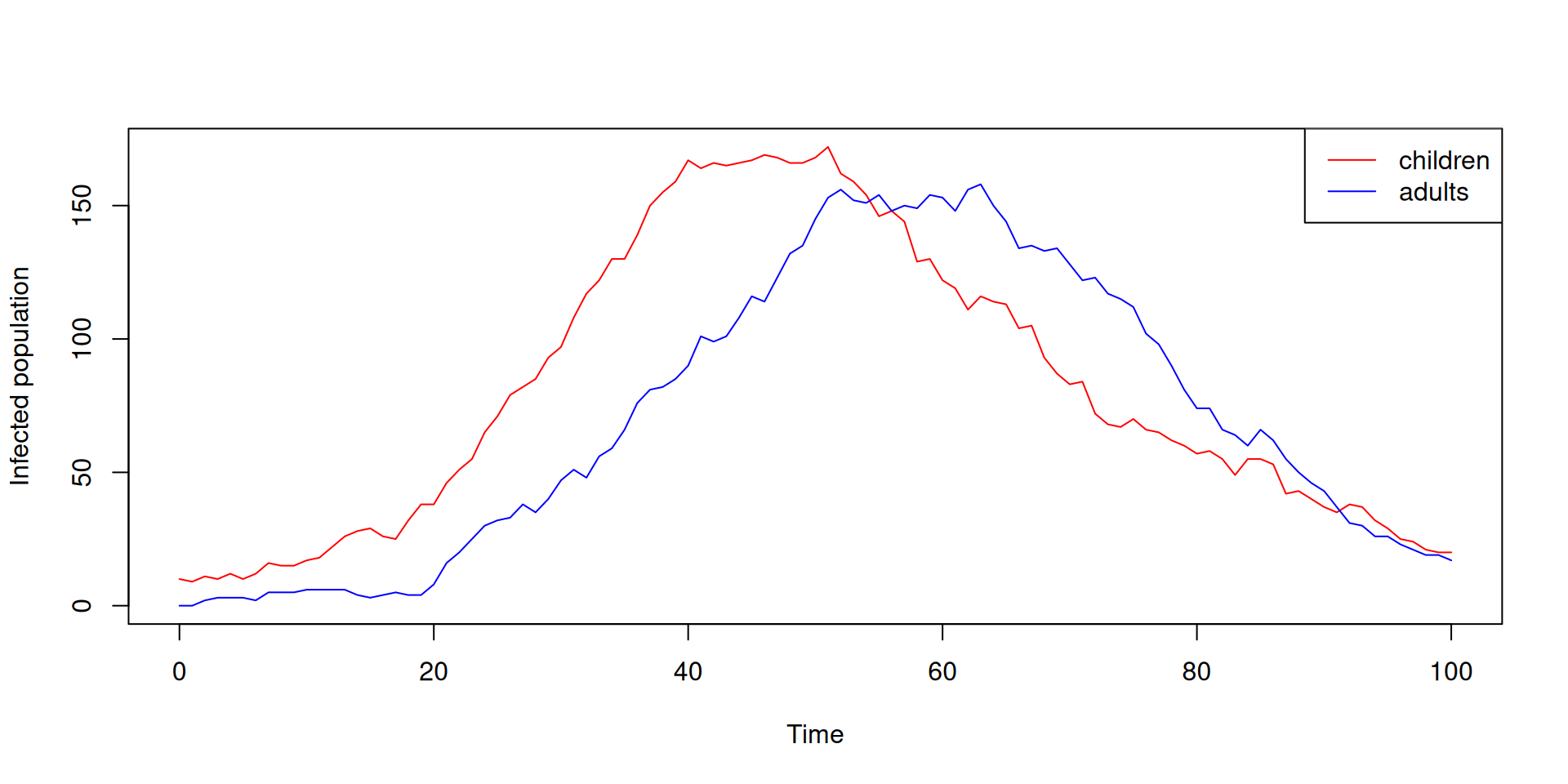
Key features of arrays
- All arrays (whether state variable or parameter) need a
dimequation - No use of
i,jetc on LHS - indexing on the LHS is implicit - Support for up to 8 dimensions, with index variables
i,j,k,l,i5,i6,i7,i8 - Functions for reducing arrays such as
sum,prod,min,max- can be applied over entire array or slices
Arrays: model with age and vaccine
sir_age_vax <- odin({
# Equations for transitions between compartments by age group
update(S[, ]) <- new_S[i, j]
update(I[, ]) <- I[i, j] + n_SI[i, j] - n_IR[i, j]
update(R[, ]) <- R[i, j] + n_IR[i, j]
update(incidence) <- incidence + sum(n_SI)
# Individual probabilities of transition:
p_SI[, ] <- 1 - exp(-rel_susceptibility[j] * lambda[i] * dt) # S to I
p_IR <- 1 - exp(-gamma * dt) # I to R
p_vax[, ] <- 1 - exp(-eta[i, j] * dt)
# Force of infection
m <- parameter() # age-structured contact matrix
s_ij[, ] <- m[i, j] * sum(I[j, ])
lambda[] <- beta * sum(s_ij[i, ])
# Draws from binomial distributions for numbers changing between
# compartments:
n_SI[, ] <- Binomial(S[i, j], p_SI[i, j])
n_IR[, ] <- Binomial(I[i, j], p_IR)
# Nested binomial draw for vaccination in S
# Assume you cannot move vaccine class and get infected in same step
n_S_vax[, ] <- Binomial(S[i, j] - n_SI[i, j], p_vax[i, j])
new_S[, 1] <- S[i, j] - n_SI[i, j] - n_S_vax[i, j] + n_S_vax[i, n_vax]
new_S[, 2:n_vax] <- S[i, j] - n_SI[i, j] - n_S_vax[i, j] + n_S_vax[i, j - 1]
initial(S[, ]) <- S0[i, j]
initial(I[, ]) <- I0[i, j]
initial(R[, ]) <- 0
initial(incidence, zero_every = 1) <- 0
# User defined parameters - default in parentheses:
S0 <- parameter()
I0 <- parameter()
beta <- parameter(0.0165)
gamma <- parameter(0.1)
eta <- parameter()
rel_susceptibility <- parameter()
# Dimensions of arrays
n_age <- parameter()
n_vax <- parameter()
dim(S, S0, n_SI, p_SI) <- c(n_age, n_vax)
dim(I, I0, n_IR) <- c(n_age, n_vax)
dim(R) <- c(n_age, n_vax)
dim(m, s_ij) <- c(n_age, n_age)
dim(lambda) <- n_age
dim(eta) <- c(n_age, n_vax)
dim(rel_susceptibility) <- c(n_vax)
dim(p_vax, n_S_vax, new_S) <- c(n_age, n_vax)
})Arrays: boundary conditions
Multiline equations can be used to deal with boundary conditions, e.g. we have
new_S[, 1] <- S[i, j] - n_SI[i, j] - n_S_vax[i, j] + n_S_vax[i, n_vax]
new_S[, 2:n_vax] <- S[i, j] - n_SI[i, j] - n_S_vax[i, j] + n_S_vax[i, j - 1]which we could also write as
new_S[, ] <- S[i, j] - n_SI[i, j] - n_S_vax[i, j]
new_S[, 1] <- new_S[i, j] + n_S_vax[i, n_vax]
new_S[, 2:n_vax] <- new_S[i, j] + n_S_vax[i, j - 1]or another way of writing this would be to use if else