Warning in odin2::odin({: Found 2 compatibility issues
Replace calls to 'user()' with 'parameter()'
✖ sd <- user()
✔ sd <- parameter()
Replace calls to r-style random number calls (e.g., 'rnorm()') with monty-stye
calls (e.g., 'Normal()')
✖ update(y) <- y + rnorm(0, sd)
✔ update(y) <- y + Normal(0, sd)Future of the tools
What is new?
- Since
odinv1 (classic odin, pre 2020)- comparison to data and likelihood support
- run multiple sets of parameters at once
- run in parallel
What is new?
- Since
odin.dust(2024 rewrite)- more efficient parameter updating
- parameter packers
- better parallelism
- periodic variable resetting (
zero_every) - better error messages
- compile time array bounds checking
- debugging support
Syntax changes
user()->parameter()- Discrete models have a proper time basis,
dtis now a reserved word - No longer use R’s names for distribution functions
- Named arguments allow clearer code
Automatic migration
You can use odin_migrate() to rewrite code.
Limitations
- Much slower compilation time (we will mitigate this by using js)
- Delays less flexible than in version 1 (cannot be used in discrete time models, the default argument has been removed)
Practical considerations
- Handful of missing features from
odinv1 anddustv1 (odin.dust)- delayed delays
- mixed time models
- compilation to JavaScript
- extendable via C++
Practical considerations
- The great package migration
dust2todustodin2toodinand all onto CRAN- Once on CRAN, our ability to change the
dustandmontyC++ code is reduced
Planned new features
GPU support
- Massively parallel stochastic models
- Proof-of-concept: 1 consumer GPU = 5-10 32-core nodes
- Simulation with many parameter sets harder
MPI/HPC support
- Alternative approach to parallelism
- based on message passing, rather than shared memory
- Use CPU-based HPC with fast networking
- We are interested in hearing about models that can take advantage of these levels of parallelism
More radical changes to the DSL?
- Support for events
- More bounds checking and debugging support
- Vector-returning functions (multinomial, matrix mutiplication, etc)
- Describe models in terms of flows
- Composable sub models (I am told this is very hard!)
- Improve monty’s little DSL!
- What else?
Improvement of supported particle methods?
- SMC^2, IF^2
- PF other than bootstrap
- Methods based on estimates of ratio of density rather than ratio of density estimates
Automatic differentiation
Gradient vs random walk
- Goal: Sample from the posterior efficiently
- 🐢 Random Walk MCMC:
- No knowledge of shape of posterior
- Can get stuck in tight or curved regions
- ⚡ Gradient-based methods:
- Use the local slope to move efficiently
- Better scaling in high dimensions
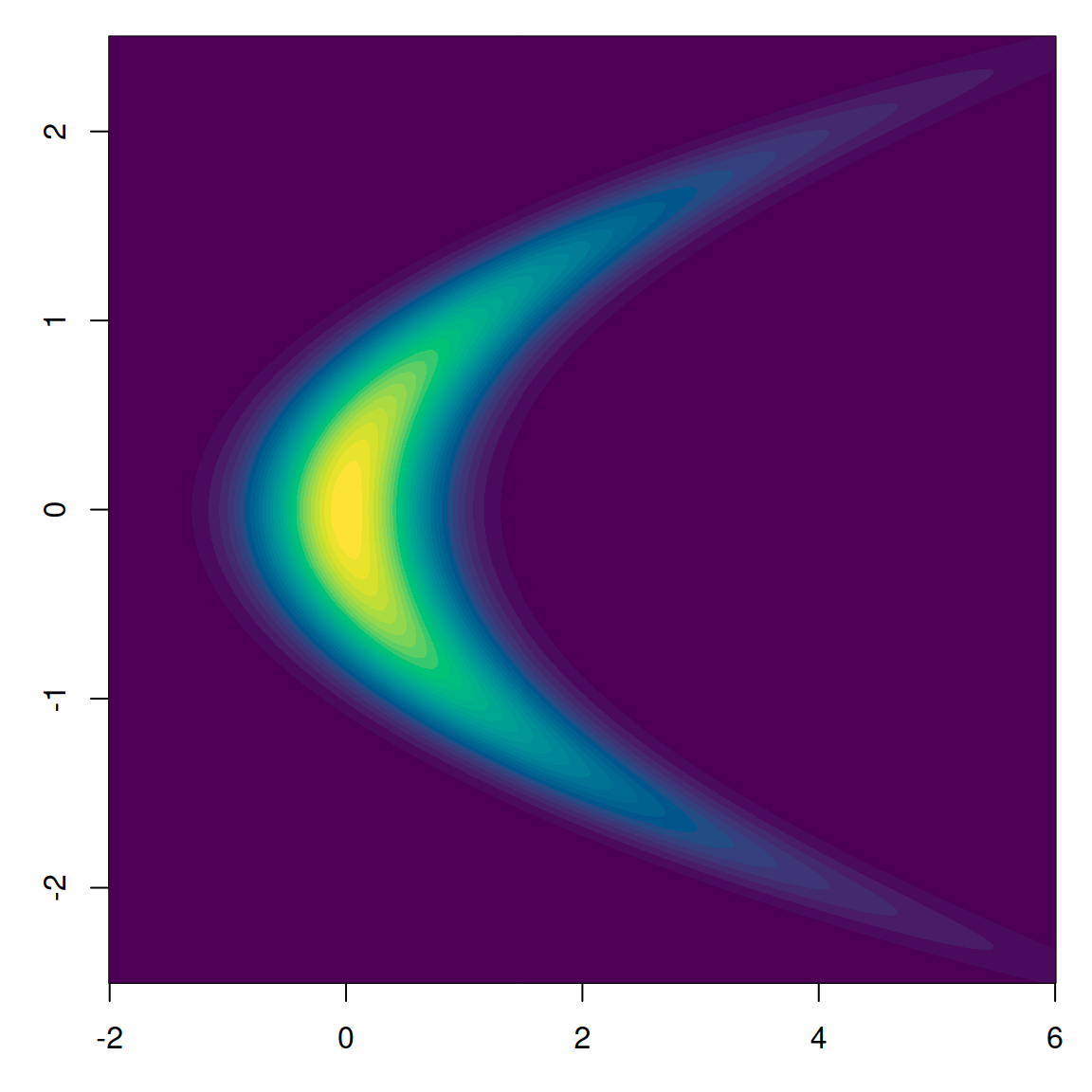
🍌 The Banana Problem
- This posterior has a strong nonlinear correlation
- Random walk proposals struggle to explore this space
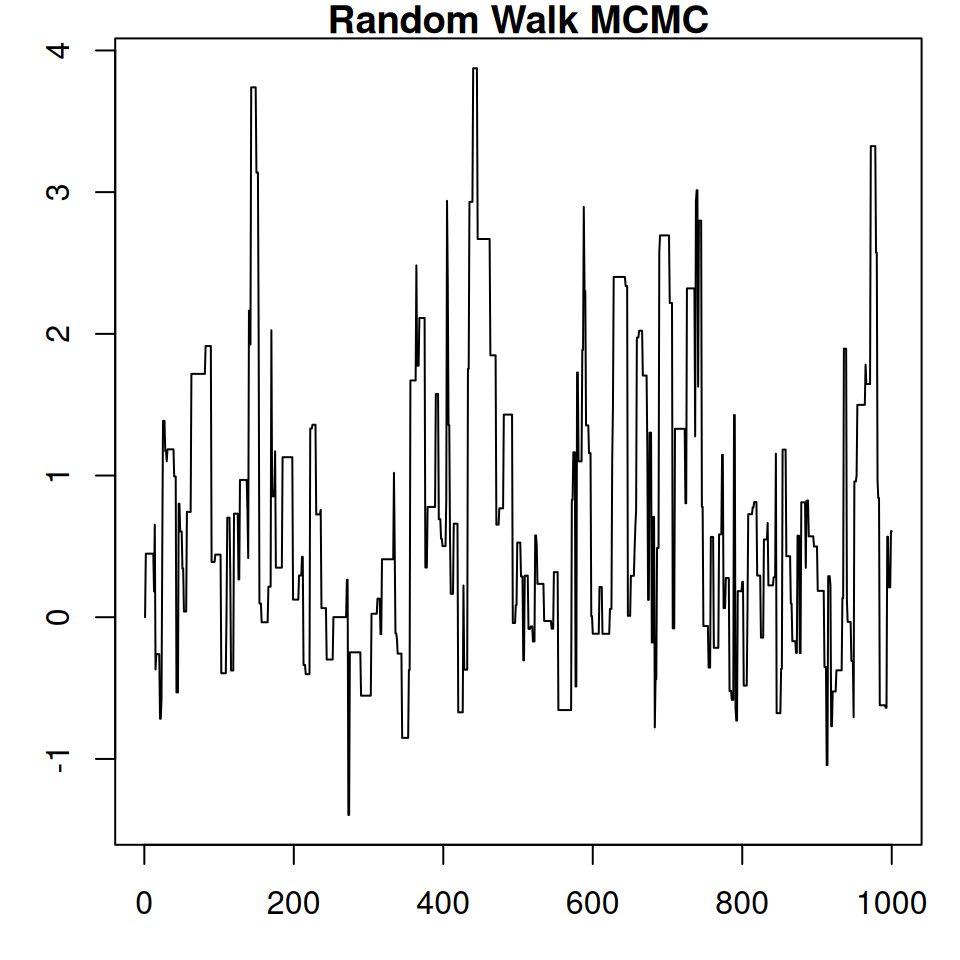
🐢 Random Walk MCMC: Limitation
set.seed(42)
sampler_rw <- monty_sampler_random_walk(vcv = diag(2)*1.5)
samples_rw <- monty_sample(m, sampler_rw, n_steps = 1000, initial = c(0,0))⡀⠀ Sampling ■ | 0% ETA: 3s✔ Sampled 1000 steps across 1 chain in 42ms- Acceptance rate 0.236
- Small steps to avoid rejection → slow mixing
- Misses curved geometry
- Inefficient in higher dimensions
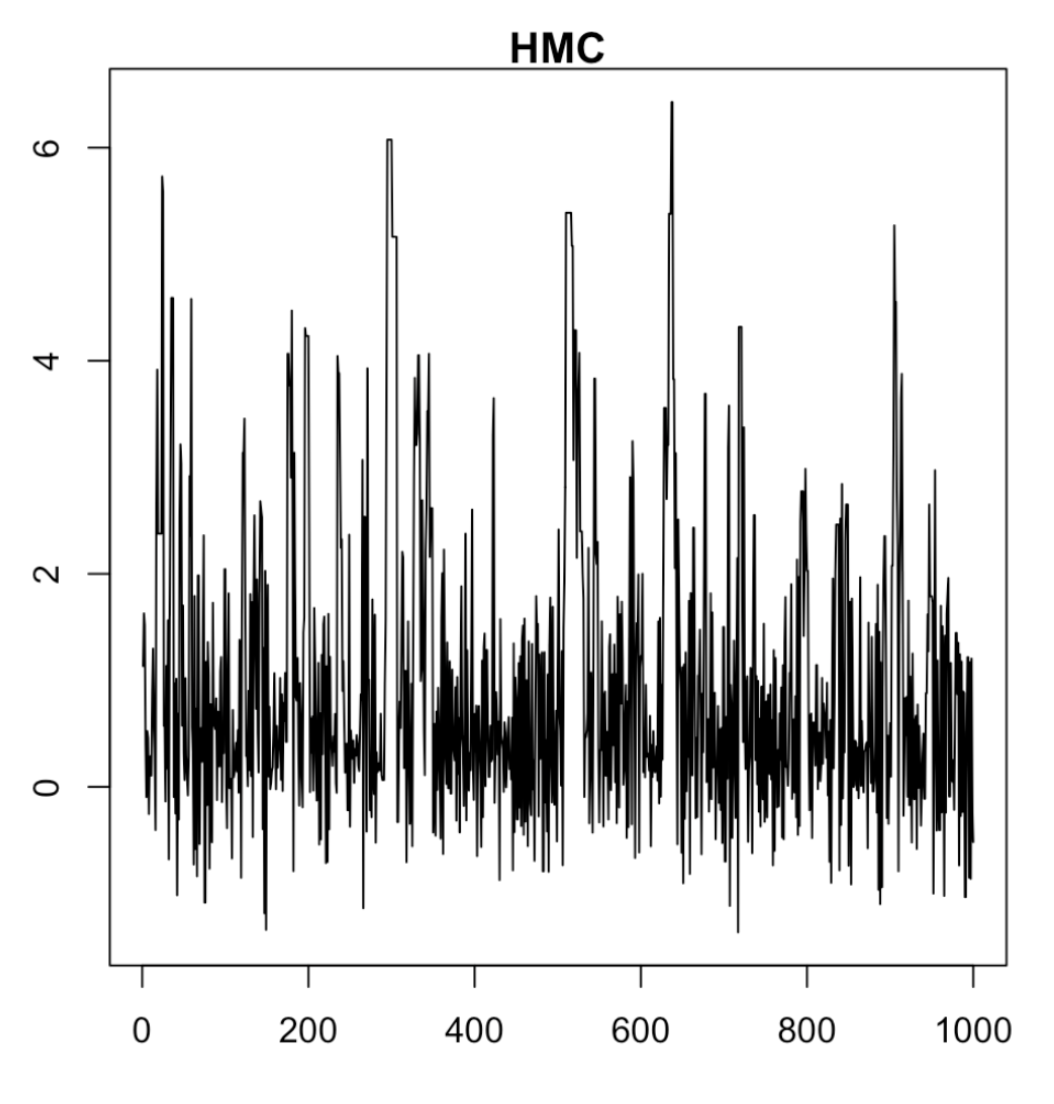
⚡ Gradient-Based: Faster & Smarter
- Acceptance rate 0.236
- Uses gradient of the log posterior
- Efficiently explores curved shapes
- Much better mixing in fewer steps
- But potentially expensive to compute gradients
Reverse AutoDiff in odin
- Think of your model as a computational graph: data + parameters → output
- Reverse AD walks backward through this graph to efficiently compute gradients
- ✅ More accurate than numerical methods
- ✅ Much faster (especially in high dimensions)
🛠 In odin, you write the model normally — gradients come for free
✅ Summary
- Gradient-based methods like HMC/NUTS:
- Are more efficient, especially for complex or high-dimensional posteriors
- Adapt to local geometry (no tuning random walk scale!)
- Often yield better convergence diagnostics
- 🚀 For users fitting models: you’ll get faster, more reliable inference with gradients when available!
🗺️ Autodiff roadmap
- Simple support implemented as a proof-of-concept
- deterministic discrete time models with no arrays
- Expand to support ODE models, models with arrays
- Fully implement algorithms in monty that can exploit gradients
- HMC, NUTS, variational inference
Parallel tempering