Malariaverse Workshop Calibration
Pete Winskill
Welcome back!
Session aims:
- Understand why we need to “calibrate” a site
- Understand when we need to calibrate/re-calibrate
- Introduction to 📦[cali] for model calibration
- Common challenges/issues when calibrating
Why calibrate?
- Given all the information about a site (vectors, season PfPr2-10 etc…). We still need to make our best estimation of the “baseline” level of transmission.
- One relatively simple way of doing this is by modifying the modelled baseline until our model output best matches some observations (e.g. PfPr2-10).
Calibration process
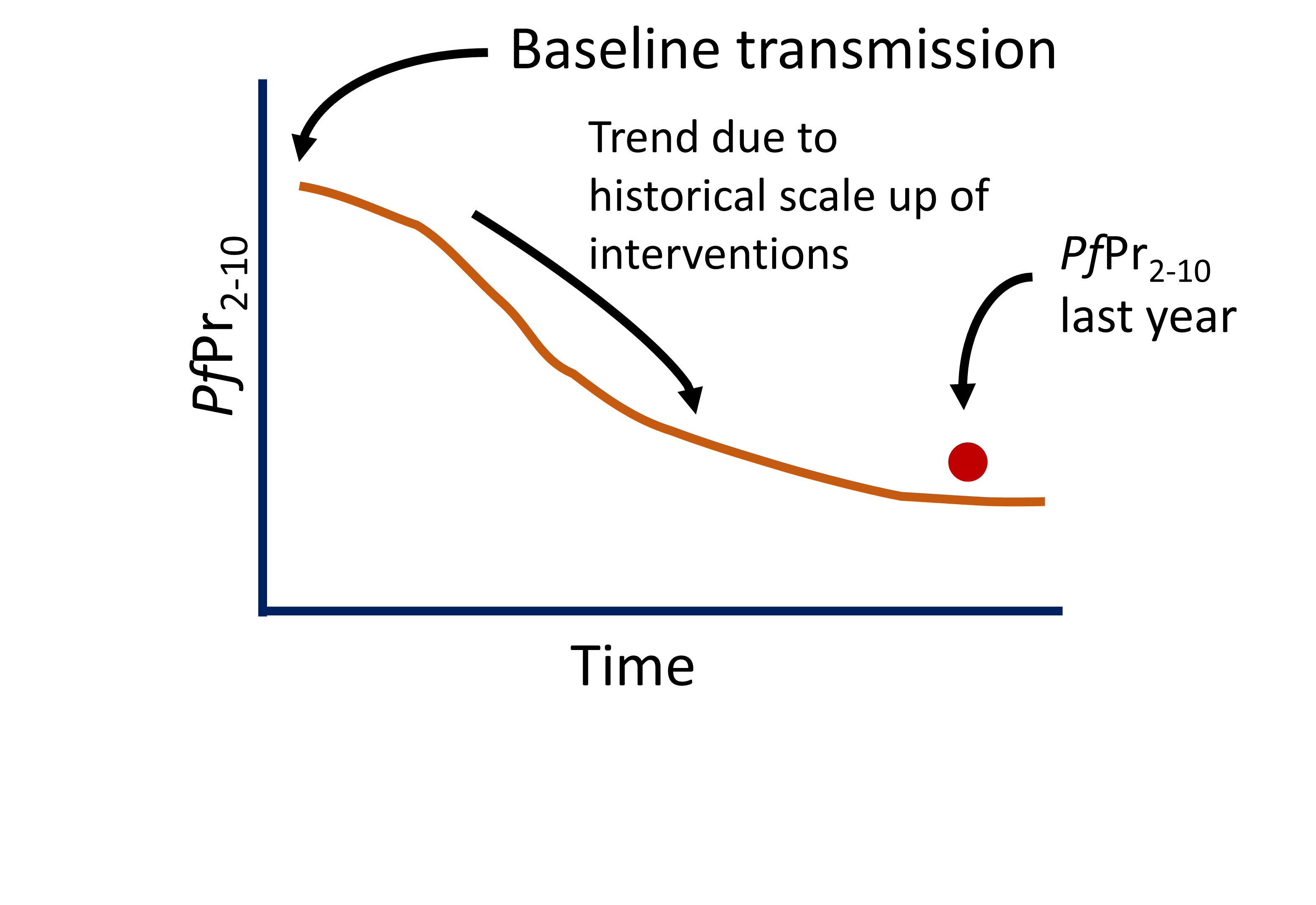
Calibration process
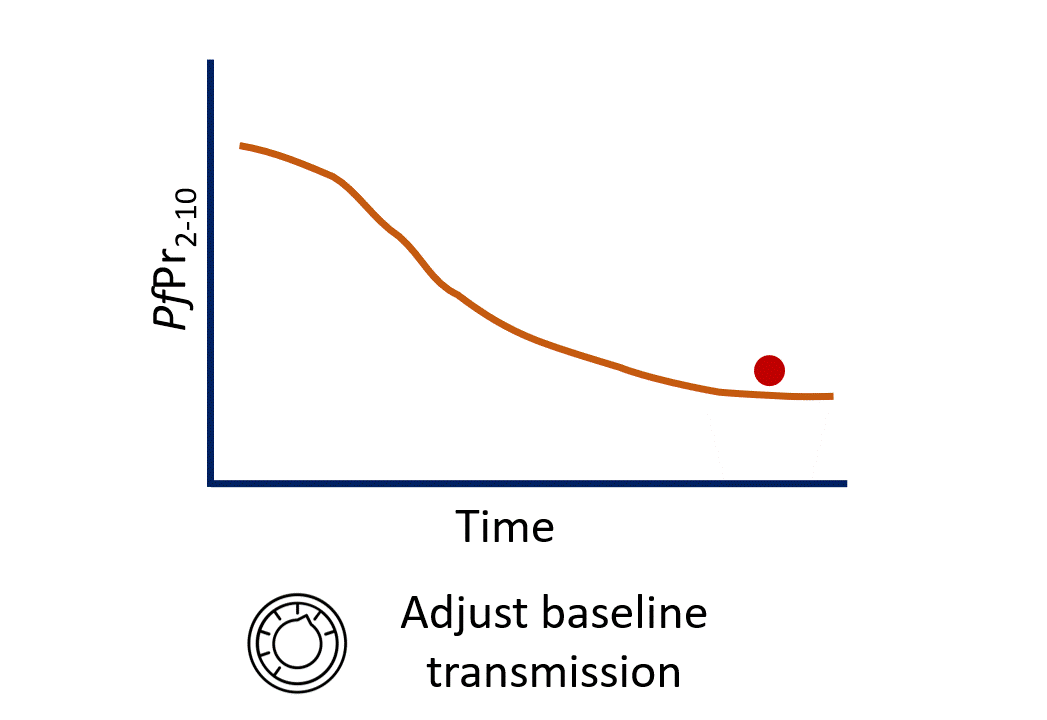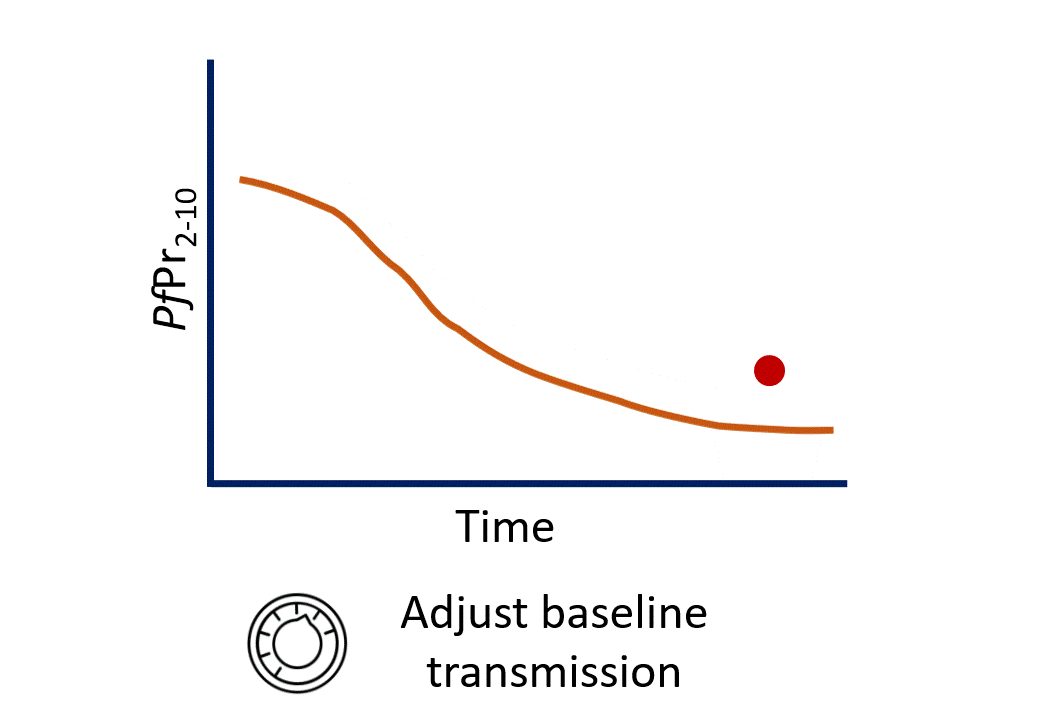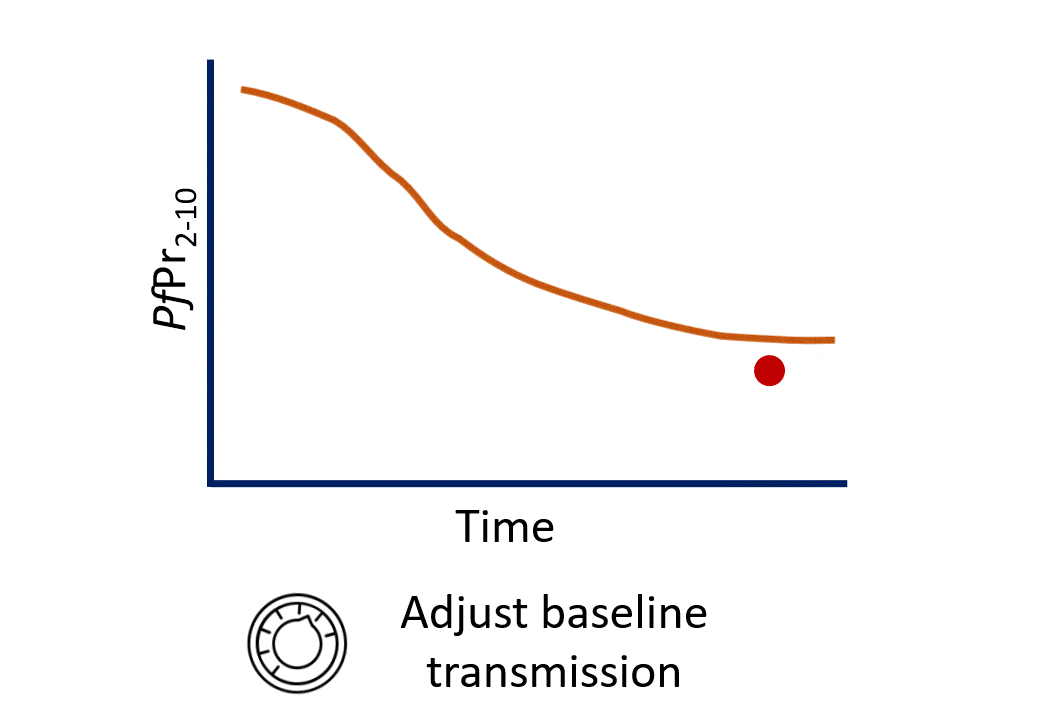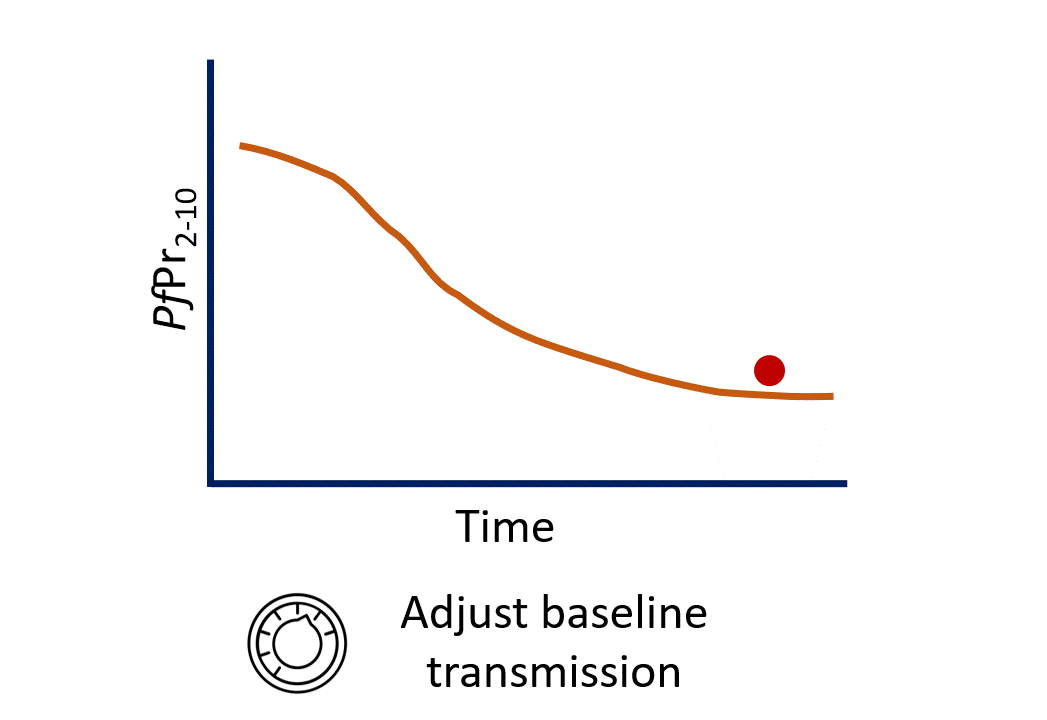
Calibration process
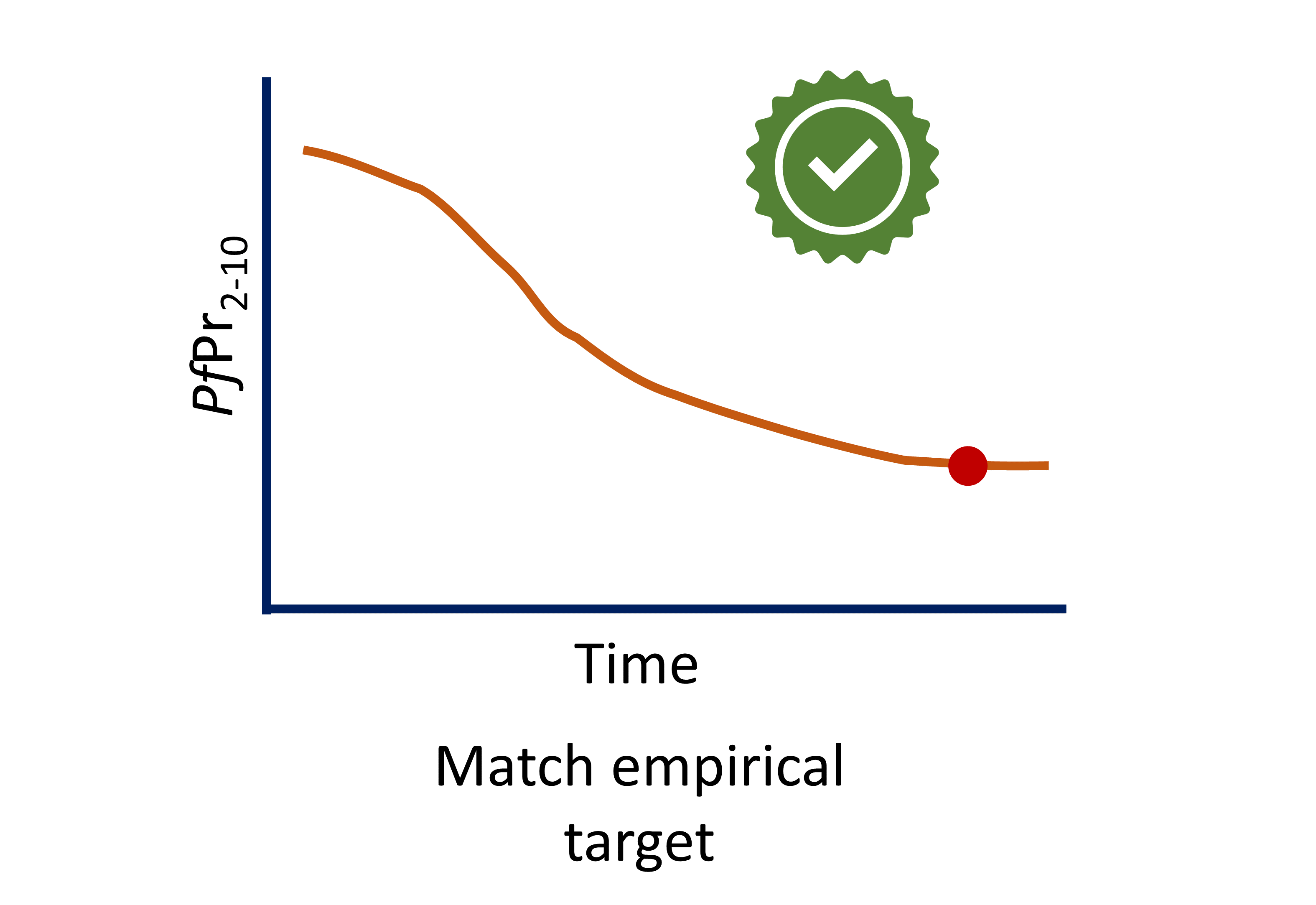
Foresite calibrations
- Site files in 📦 [foresite] have been calibrated
$eir - The EIR (Entomological Inoculation Rate) is our “dial” we can turn to change baseline transmission.
- Turning the EIR dial is essentially leading to changes in the carrying capacity of the environment we are modelling - how many mosquitoes it can support.
Calibration notes
- Data 📑
- It will be your decision on what data (and how much of it) to calibrate to
- It is important to understand how and when data were collected, for example data collected at peak malaria season may look very different to that collected in the low season.
- Warmup 💪
- 📦 malariasimulation runs do not automatically run a warmup period which is necessary for the model to fully equilibrate. In general it is always advisable to add a warmup period to your simulation runs. Some outputs are more sensitive than others and exploration of a suitable warmup period to use should be performed prior to calibration.
- Subsequent model runs after calibration should use the same warmup period as the calibration.
When to calibrate
- Calibration or re-calibration is not always required. In general, you will need to calibrate if:
- You have created a new site file
- You have modified any site inputs that occur before or at the same time as the data you are trying to calibrate to
- The data you are calibrating to has changed
- There has been a change to an underlying assumption that would impact the disease dynamics in a model run
Cali
- We can use 📦cali for model calibration.
- The
calibrate()function takes atarget, for example annual PfPr2-10 for the years 2015-2020. - The user defines a
summary_function, a function that takes the raw model output and produces the model estimate corresponding to thetarget. In this case our summary function will summarise the model-estimated PfPr2-10 for the years 2015-2020 calibrate()will then search a range of EIRs until the outputs ofsummary_functionare within a user-defined tolerance of thetarget
Cali
- It is always good practice to run a simulation and test your
summary_functionbefore using it incalibrate()- this will help with debugging 🐞. - There a lots of other tuning and model options, so please see
?calibrate()for more detailed information.
Challenges
- 📦cali is (hopefully) convenient, however it doesn’t remove the need to run the model multiple time when calibrating. Therefore be prepared for the process to be lengthy, often requiring calibration runs to be performed on the high performance cluster.
Challenges
- Available data and model outputs are not guaranteed to play nicely

Challenges
Sometimes calibration can look good:
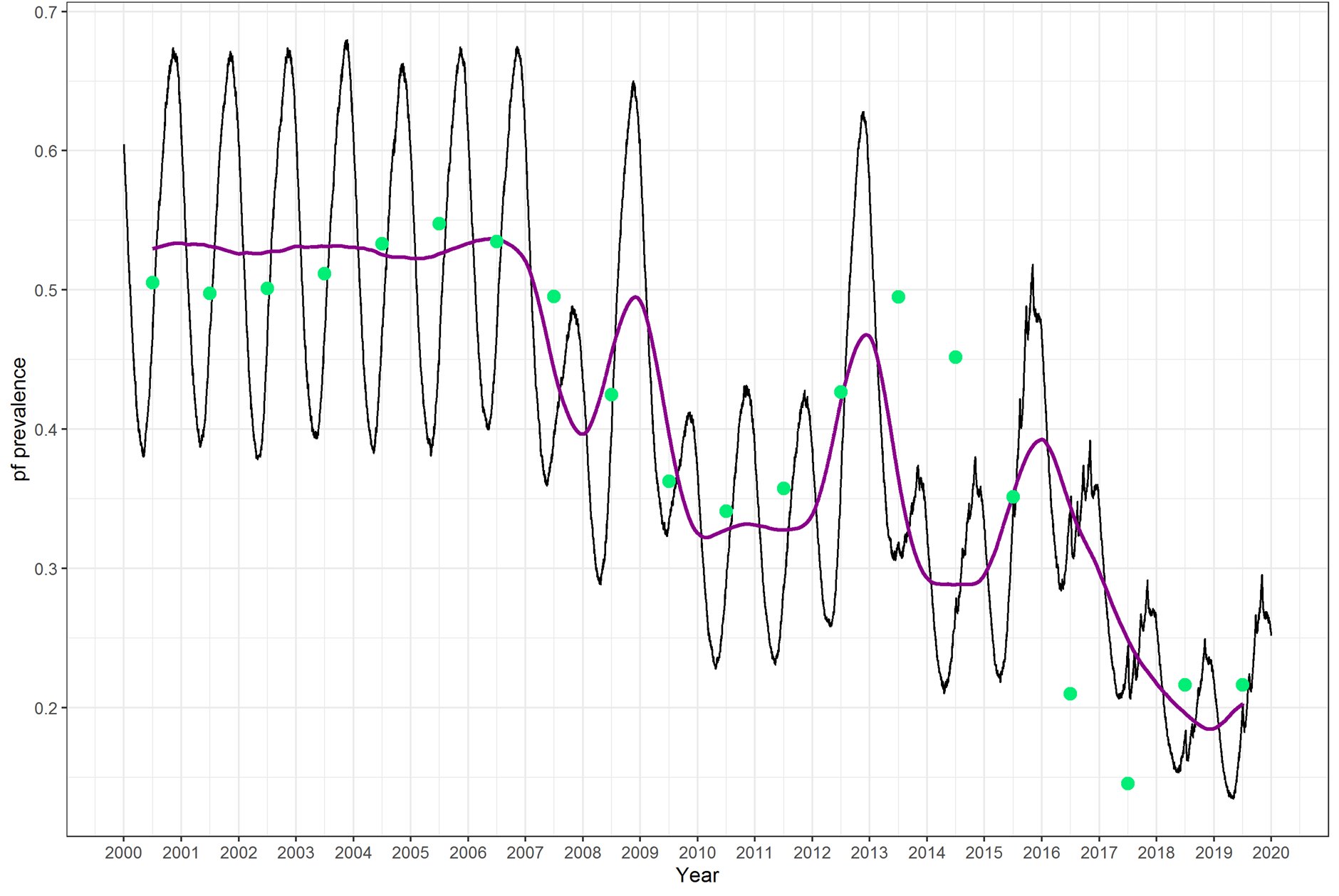
Challenges
…and sometimes not 😢 :
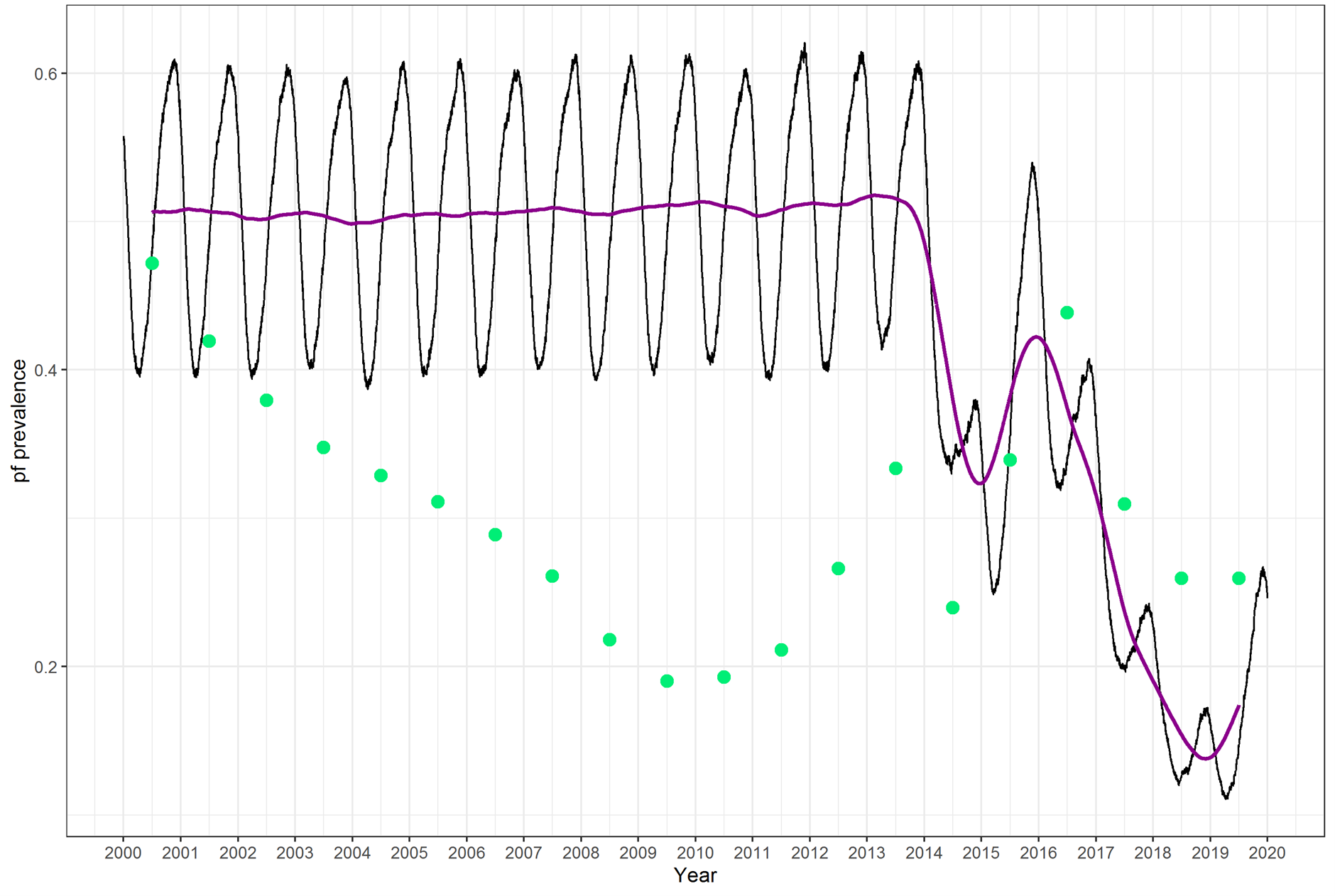
Challenges
- Calibration can be difficult when results are dominated by stochasticity:
- low transmission
- small populations
- calibrating to a sub-group in the population
- etc…
Let’s give it a try!
- Run
mvw::run_tutorial("Calibration")

